Innovative Techniques in Protein Design: A Look into PLAID
This article explores the novel approaches in protein generation using latent diffusion models, particularly focusing on the PLAID framework that promises substantial advances in protein design and structure prediction.
By · 2025-04-08 15:30:00 · From The Berkeley Artificial Intelligence Research Blog via bair.berkeley.edu
The recent awarding of the 2024 Nobel Prize to AlphaFold2 marks a pivotal moment for the intersection of artificial intelligence (AI) and biology, highlighting the role of AI in understanding protein structures. The current landscape presents additional questions: What comes next after protein folding? How can we leverage these advancements to generate new proteins effectively?
Core Idea/Problem
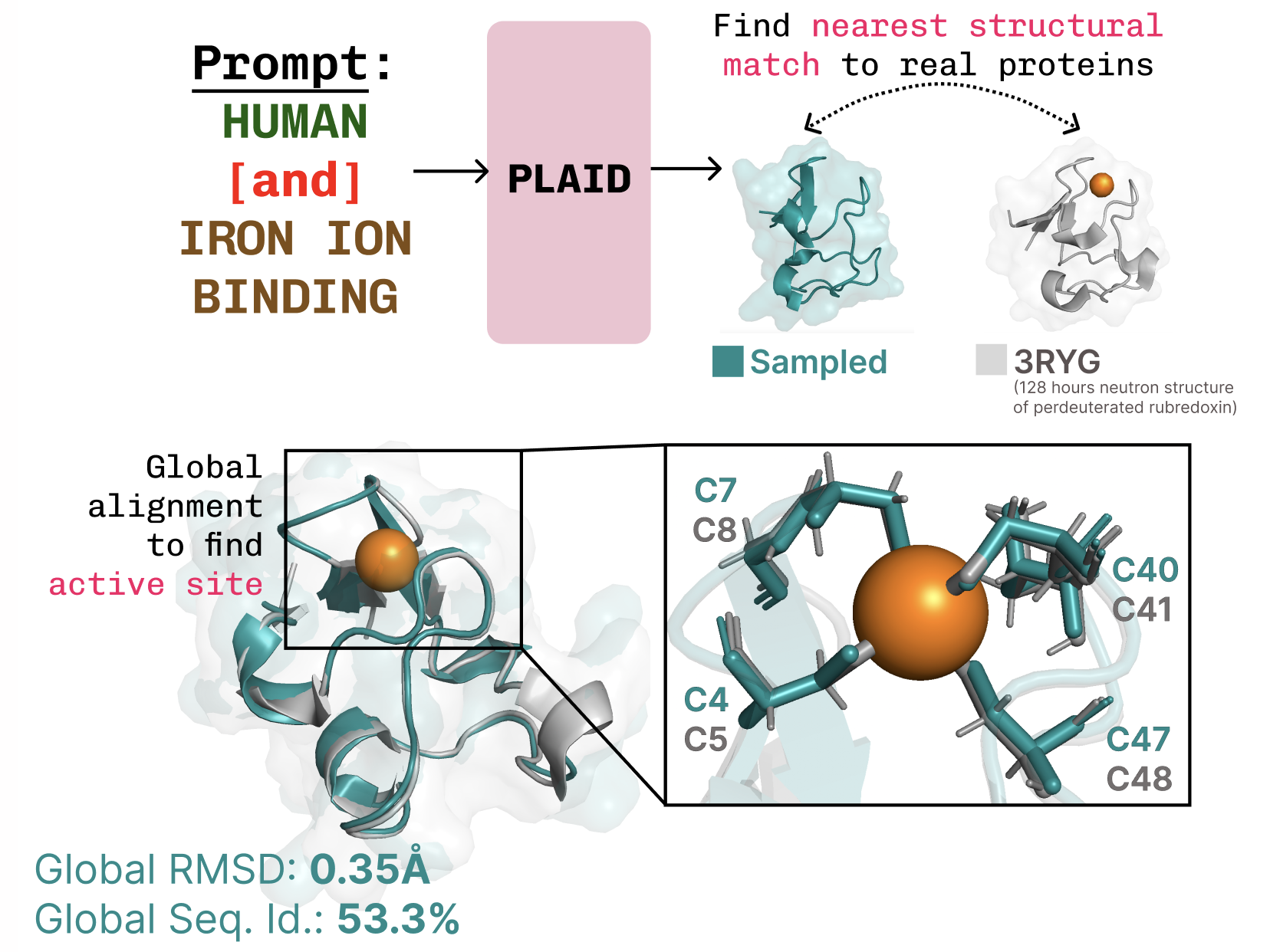
The work surrounding PLAID (a multimodal generative model) focuses on generating both the one-dimensional sequence and three-dimensional structure of proteins simultaneously. This model learns from the latent space of existing protein folding algorithms and accepts prompts based on compositional functions and organisms. Its development indicates a significant advancement over traditional models, which typically only generate one dimensional data or rely heavily on more limited structural databases.
What the Data/Details Show
In PLAID, researchers created a system that can be trained on sequence databases that are several orders of magnitude larger than typical structure databases. This addresses the limitation of previous models that could only partially generate proteins. Notably, PLAID grapples with the multimodal co-generation problem by producing discrete sequences alongside continuous all-atom structural coordinates.
From Structure Prediction to Real-World Drug Design
While previous diffusion models have shown promise, they often encounter limitations that hinder their practical use in drug design and bioengineering. Key issues include:
- All-atom generation: Many existing algorithms produce only backbone atoms. A complete all-atom structure requires knowledge of the sequence, posing a significant complication.
- Organism specificity: Proteins intended for human therapeutic applications must be “humanized” to prevent destruction by the immune system.
- Control specification: The complexity of drug design necessitates precise control over variables, such as the format of delivery systems (e.g., deciding between tablets and vials).
Generating “Useful” Proteins
Simply generating proteins is an incomplete goal; instead, it is critical to have control over the generation process to yield useful proteins. The PLAID model aims to provide an interface for this control, potentially mirroring techniques used in image generation, where users specify functions and traits creatively.

Interface concepts for controlling protein generation in a manner similar to compositional textual prompts in image generation.
In this venture, PLAID aims to enhance user interaction through a text-based interface for two primary axes: function and organism.
Training Using Sequence-Only Training Data
A major advantage of the PLAID framework lies in its requirement to train solely on sequence data. This opens up significant access to broader databases, enabling the model to leverage considerably larger datasets which are often more readily available compared to structural data.
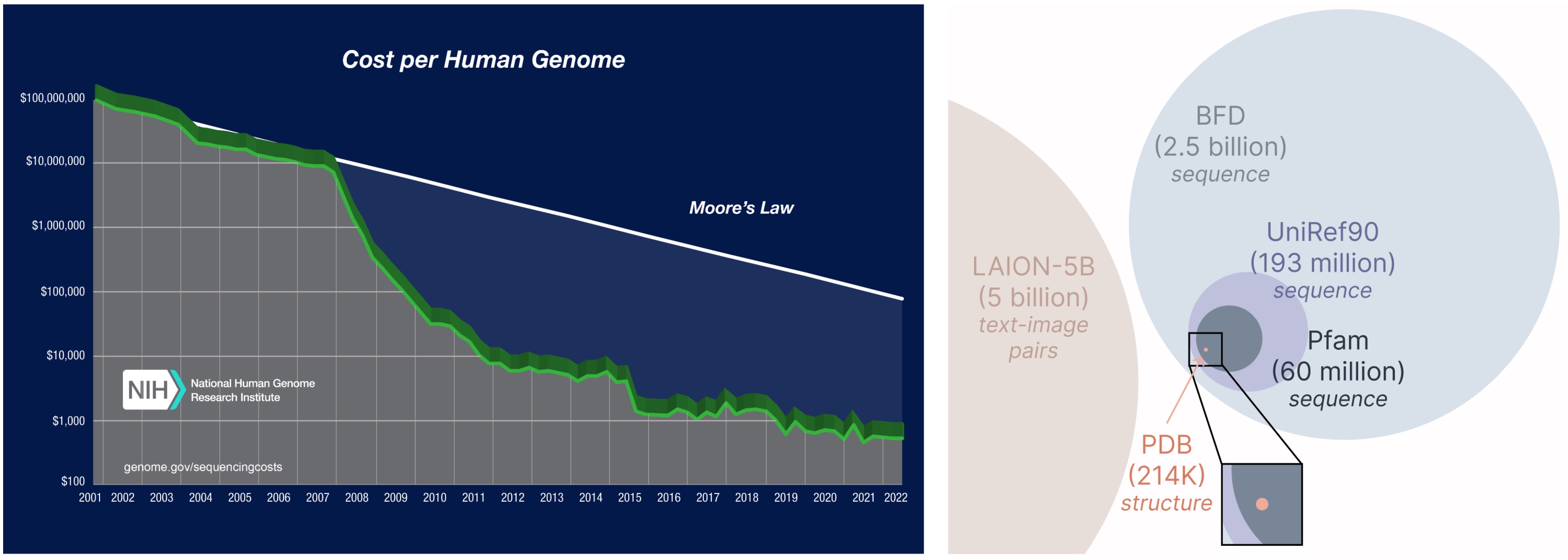
The significant cost disparity in obtaining protein sequences versus experimentally determining their structure.
How It Works / The Mechanism
The PLAID model operates by learning a diffusion model over the latent space of a protein folding model. During training, generators only need sequence data, but they output both sequence and structural embeddings during inference. The ESMFold model, an advancement over AlphaFold2, provides the necessary framework by integrating a protein language model for decoding structures effectively.
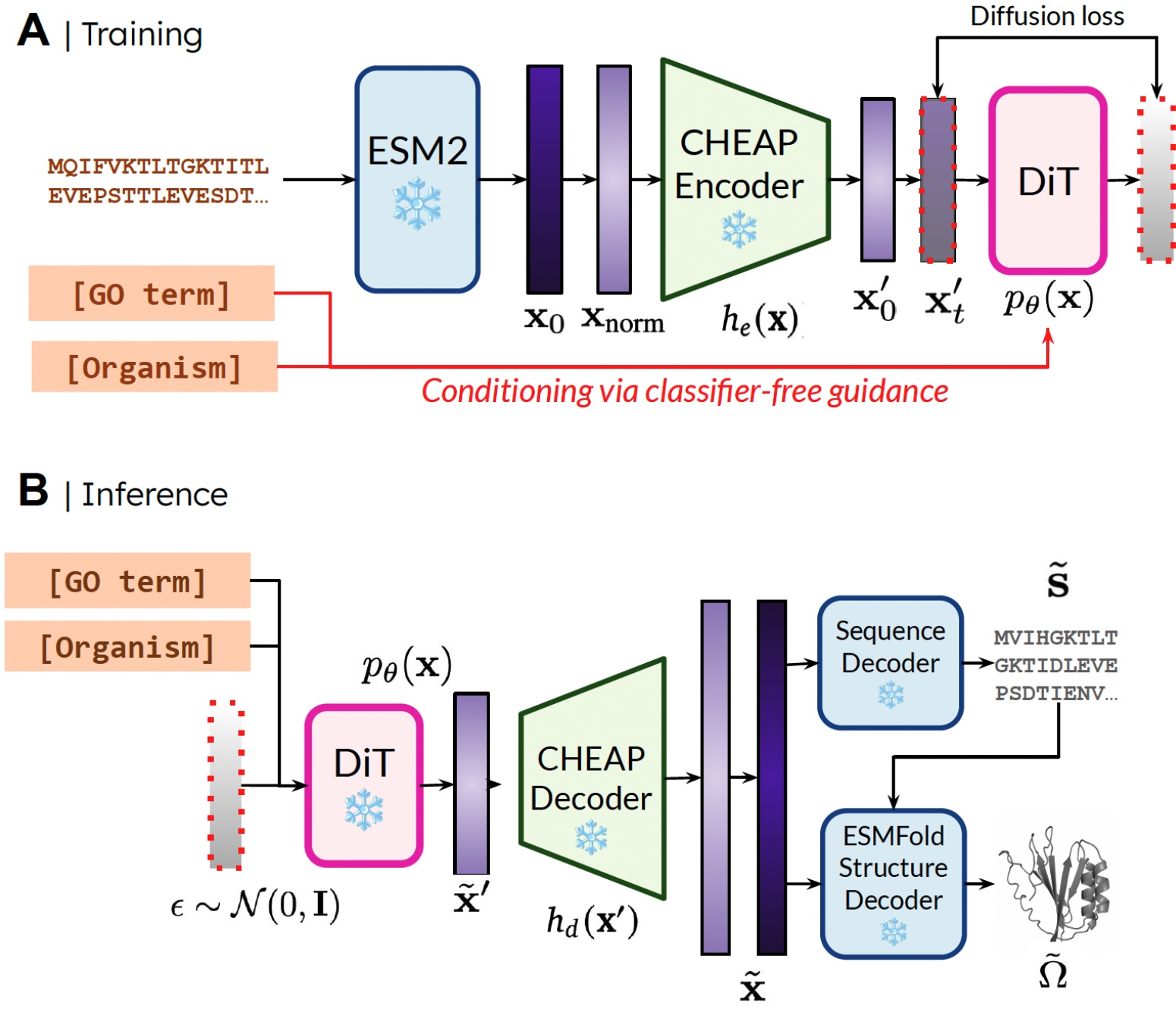
Training and inference flow in the PLAID model.
This architecture allows it to use previously acquired structural knowledge in the protein design task, drawing parallels to how vision-language models in robotics are employed to facilitate complex tasks.
Implications & Use Cases
The implications of this work extend across various fields. Here are a few use cases:
- Pharmaceutical Development: By controlling protein properties, PLAID can assist in designing drugs that are more effectively tailored to user needs.
- Biotechnology Applications: Custom-designed proteins could revolutionize everything from enzyme development to agricultural applications, ensuring that bioengineered organisms perform optimally in their intended environments.
- Basic Research: Researchers could utilize PLAID to generate specific proteins required for experimental setups, vastly accelerating laboratory workflows.
This model underscores a shift towards more intuitive and effective processes in biological research and engineering.
Limitations, Caveats & Unknowns
Despite its innovative framework, there are constraints to PLAID’s current capabilities:
- The reliance on existing protein folding models and their inherent assumptions can pose limitations on the types of proteins that can be effectively generated.
- As with any generative model, validation through empirical methods is essential to ensure that generated proteins function as intended, particularly in therapeutic contexts.
- The complexities of human immune response and protein interactions add further challenges beyond mere sequence generation.
What’s Next
The potential for PLAID extends into the realm of multimodal generation that could influence broader biological systems. As advancements continue, researchers might adapt PLAID methods to tackle more complex systems, including those that intersect with nucleic acids and molecular ligands.
This exciting work marks a step forward in protein design, signaling a readiness for scalable and efficient generation methods in various biotechnological fields. Interested researchers are encouraged to collaborate in expanding PLAID’s capabilities and exploring its applications in real-world settings.
#Repurposing #Protein #Folding #Models #Generation #Latent #Diffusion #Berkeley #Artificial #Intelligence #Research #Blog
Slug: /repurposing-protein-folding-models-for-generation-with-latent-diffusion-the-berkeley-artificial-intelligence-research-blog