Identification of BCR-Related Genes Using WGCNA
Understanding WGCNA: A Systems Biology Approach
Weighted Gene Co-Expression Network Analysis (WGCNA) is a powerful systems biology method that provides insights into gene association patterns across various samples. This approach excels in identifying sets of genes that change in a coordinated fashion, thus helping researchers pinpoint candidate genes based on their interconnectivity. Not only can WGCNA reveal associations with phenotypes, but it also enables the identification of potential marker genes or therapeutic targets.
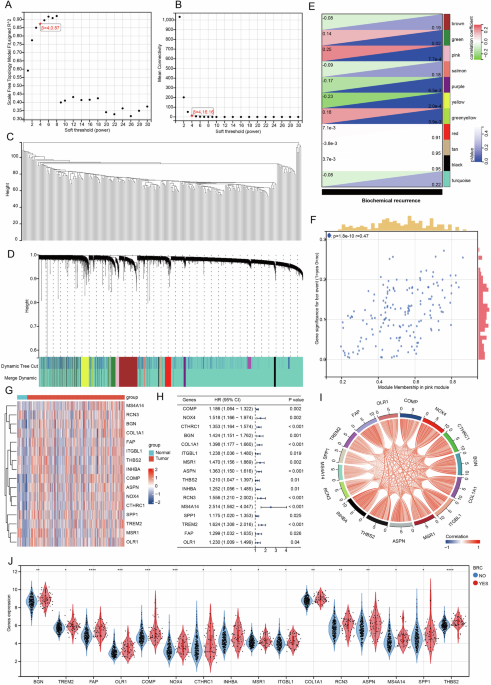
In our study, we utilized WGCNA to explore regulatory genes linked to biochemical recurrence (BCR) in prostate cancer (PRAD), using a dataset from GSE116918 that comprises 248 samples. The initial step involved screening to determine the optimal soft threshold, followed by clustering analysis of the samples. This led to the identification of 11 stable modules, with the pink module demonstrating the strongest correlation with BCR.
Within this pink module, we identified 162 genes, out of which 16 were highly expressed in PRAD and related to patient progression-free intervals (PFI). Notably, these genes showed a positive correlation in the TCGA-PRAD dataset, further validating their relevance to BCR. The analysis revealed that the expression levels of these 16 genes were significantly higher in patients experiencing BCR compared to those without.
Functional Analysis of BCR-Associated Genes
To decode the functions of the identified 16 BCR-associated genes, we turned to the KEGG and GO databases. The KEGG analysis linked these genes to several important biological pathways, including:
- ECM-receptor interaction
- Phagosome formation
- Focal adhesion
- PI3K-Akt signaling pathway
The GO analysis provided additional insights, indicating that these genes mainly regulate:
- B-cell differentiation
- Autophagic cell death
- Macrophage differentiation
Further exploration through the GSCA database highlighted correlations with essential processes such as cell cycle activation, epithelial-mesenchymal transition (EMT), and immune regulation pathways. An examination of copy number variations among these genes disclosed significant alterations in several, including CTHRC1, FAP, and THBS2.
We also observed that these genes exhibited higher expression levels in advanced pathological stages, including higher T stages, N stages, and Gleason scores, underscoring their potential role in tumor progression.
Cluster Analysis Based on BCR-Related Genes
The next phase involved clustering TCGA-PRAD samples utilizing the Negative Matrix Factorization (NMF) algorithm. To determine the most suitable method for subgroup categorization, we assessed co-expression curves. Our analysis revealed that the optimal division was into two clusters. The clustering heat map illustrated ample concentration in color distribution, signifying distinct groups.
Interestingly, patients in cluster 1 indicated a significantly better prognosis compared to cluster 2. Further evaluation of BCR-associated gene expression across these clusters showed noteworthy differences. Our analysis across multiple pathological parameters reinforced this observation, revealing significant variances in patient distributions across various T stages, N stages, PSA levels, and Gleason scores.
Correlating BCR-Related Genes with Immune Infiltration and Chemosensitivity
Using the XCELL algorithm, we quantified immune cell infiltration across TCGA-PRAD samples. The level of various immune cells, including myeloid dendritic cells and T-cells, displayed significant discrepancies between clusters. The heat map we created complemented these findings, visually portraying the immune landscape.
In our search for chemotherapeutic implications, we identified differences in IC50 values for several therapeutic agents. Notably, bicalutamide exhibited significant variability in sensitivity between the clusters, suggesting a possible stratified response to treatment.
To delve deeper, we conducted gene enrichment analysis focusing on cluster 2, revealing significant enrichment in pathways related to WNT, PI3K-AKT, and immune response, which could inform future therapeutic strategies.
Construction of a Diagnostic Model Based on BCR-Related Genes
To further explore the potential of our identified genes in diagnosing PRAD, we employed Receiver Operating Characteristic (ROC) curves. Our findings evidenced that specific genes possess robust diagnostic capabilities. We aimed to develop diagnostic models utilizing training datasets from TCGA-PRAD and validation cohorts from GSE datasets.
After rigorous testing of 108 algorithm combinations, the LASSO + LDA algorithm emerged as the most effective strategy. The area under the curve (AUC) for the training set was 0.911, indicating exceptional diagnostic potential, while validation cohorts yielded solid AUC values, validating the model.
Within our diagnostic model, 13 BCR-associated genes played crucial roles, including ASPN, CTHRC1, and TREM2, providing a foundation for clinical application.
Constructing a Prognostic Model for BCR
To forge a prognostic model, we gathered clinical data from BCR patients within the TCGA-PRAD dataset, defining the timeline from diagnosis to BCR. After merging datasets and applying random seed divisions, we screened for prognosis-related genes using univariate Cox analysis.
Six significant genes were identified, leading to the construction of a multivariate Cox model, after additional refinement through LASSO regression. Our model demonstrated reliable predictive capabilities for BCR-related prognosis, marking patients at high risk as having significantly poorer outcomes compared to those classified as low-risk.
Machine Learning to Identify Key BCR Regulatory Genes
To pinpoint key regulatory genes associated with BCR, we employed the XGBoost algorithm, interpreting results with SHAP values. This revelation presented us with the top 15 genes linked to BCR, with COMP emerging as a primary candidate.
Upon dividing the samples based on COMP expression, we discovered a significant correlation with immune cell infiltration levels. Our investigation into drug interactions revealed high binding affinities between COMP and clinically significant prostate cancer treatments. Enrichment analysis linked COMP to essential immunotherapy pathways, underscoring its potential role in therapeutic contexts.
Expression Analysis of COMP in PRAD
Our investigation extended to the expression levels of COMP, where we compared 60 PRAD samples to normal tissues using immunohistochemical staining. Results indicated that COMP is substantially overexpressed in PRAD tissues, particularly in recurrent forms compared to non-recurrent samples.
ROC analysis confirmed COMP’s predictive value regarding diagnosis and recurrence duration, presenting a compelling case for its application in clinical settings.
Inhibiting COMP Expression Suppresses Tumor Progression
Finally, we explored the functional impact of COMP via its knockdown in PCa cells. Results indicated significant reductions in cell proliferation and invasive potential. In vivo modeling in nude mice reinforced these findings, where COMP knockdown led to suppressed tumor growth and metastasis. Immunohistochemical stains demonstrated decreased proliferation markers within COMP-knockdown tumors.
This body of work not only highlights the multifaceted role of COMP in PRAD but also opens pathways for its exploration as a diagnostic and therapeutic target in clinical oncology.