Exploring RareNet: An Insight into Datasets and Methodology for Rare Cancer Analysis
Introduction to RareNet’s Evaluation Datasets
RareNet, an innovative framework for rare cancer classification, has undergone rigorous assessment with three distinct datasets. By leveraging diverse data sources, RareNet aims to enhance its generalization abilities on unseen data, thus expanding its applicability in rare cancer research. Let’s take a closer look at the datasets used in this evaluation.
The TCGA Dataset
The TCGA dataset (The Cancer Genome Atlas) is a comprehensive collection of DNA methylation data encompassing 13,325 samples across 33 different cancer types. The dataset also includes a “Normal” class, which is vital for establishing baseline comparisons between cancerous and non-cancerous tissues.
Among the 33 cancer types represented in the TCGA dataset are well-known conditions such as:
- Adrenocortical carcinoma (ACC)
- Breast invasive carcinoma (BRCA)
- Glioblastoma multiforme (GBM)
- Lung adenocarcinoma (LUAD)
The meaningful insights drawn from these diverse cancer types provide a robust foundation for genomic analyses, enhancing the reliability and accuracy of cancer diagnostics.
Importantly, the samples used in the “Normal” class originate from the same patient cohorts, which supports the validity of the comparative genomic studies conducted through RareNet.
The TARGET Dataset
In contrast to TCGA, the TARGET dataset (Tumor Alterations Relevant for Genomic-driven Therapy) focuses specifically on 5 rare cancers:
- Wilms Tumor (WT): 11 samples
- Clear Cell Sarcoma of the Kidney (CCSK): 86 samples
- Osteosarcoma (OST): 171 samples
- Neuroblastoma (NB): 221 samples
- Acute Myeloid Leukemia (AML): 130 samples
In total, the dataset comprises 777 DNA methylation samples. Notably, the corresponding “healthy” samples, categorized as normal class, consist of 158 samples. The selection of these rare cancers aligns perfectly with the training data of CancerNet, providing a cohesive framework for comparison and analysis.
The NCBI GEO Dataset
The NCBI GEO dataset taps into another rich source of methylation data, featuring samples from various rare cancers:
- Neuroblastoma: 31 samples
- Clear Cell Sarcoma of the Kidney (CCSK): 55 samples
- Acute Myeloid Leukemia (AML): 73 samples
- Normal samples: 29 samples
Altogether, this dataset comprises 188 DNA methylation samples, associated with the accession numbers: GSE54719, GSE113501, GSE125645, GSE59157, GSE62298, and GSE58477. This broad array of data aids in robust statistical modeling and the evaluation of RareNet’s performance.
Each dataset was systematically divided into training, validation, and testing sets, following the ratio of 80%-10%-10%. This method ensures that the model is thoroughly evaluated, affording insights into its real-world applicability.
The Variational Autoencoder Methodology
Given the intricacies involved in cancer classification and dimensionality reduction, a Variational Autoencoder (VAE) was chosen as a primary tool within RareNet.
What is a Variational Autoencoder?
The VAE employs an encoder to compress vast input data into a more manageable latent space, followed by a decoder to reconstruct this latent space into an output that closely resembles the original data. This dual-phase approach preserves vital information while facilitating data usability.
Data Preprocessing in RareNet
RareNet adopts a Methyldata preprocessing methodology akin to that utilized in CancerNet. This involves using a CpG density clustering technique:
- Exclusion of Non-associated CpGs: CpGs not linked to CpG islands are excluded.
- Proximity Clustering: Remaining data is assessed for Illumina 450K probes located within 100 base pairs of each other, forming cohesive clusters.
- Cluster Size Refinement: Clusters with fewer than 3 CpGs are deemed insignificant and removed.
This processing yields 24,565 clusters, with the averaged CpG (beta) values serving as input features. The VAE structure employs these features to create a 100-dimensional latent space embedding.
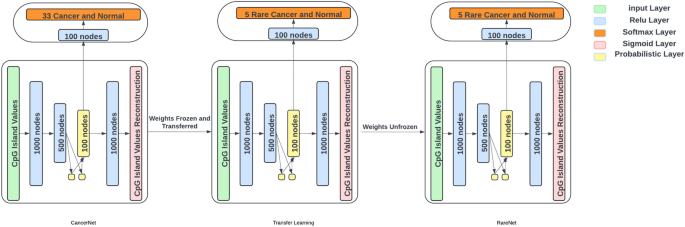
Transfer Learning and Performance Evaluation
RareNet employs transfer learning to implement insights gleaned from CancerNet, a model adept in detecting 34 common cancers. A noteworthy aspect of this approach is the use of pre-trained weights from CancerNet while keeping the encoder and decoder parameters fixed. This allows for effective adaptation to the new model’s specific requirements without altering the latent space.
Ten-fold Cross-Validation Strategy
To robustly evaluate performance, a ten-fold cross-validation strategy is employed. This approach entails dividing the dataset into ten subsets, holding one for testing while utilizing the others for model training. This systematic scrutiny enables the adjustment of model parameters, optimizing its performance and ensuring generalizability.
Architectural Specifications
The architecture of RareNet mirrors that of CancerNet, albeit tailored to focus on rare cancers, with its classifier having 6 output nodes—5 for rare cancers and 1 for normal samples. The classifier undergoes training with frozen weights from the encoder and decoder layers, facilitating a streamlined learning process.
Comparative Assessment of RareNet
To contextualize RareNet’s performance, comparative assessments against other machine learning models were executed, utilizing the Scikit-Learn Python library. Models such as Random Forest, K Nearest Neighbors, Decision Tree Classifier, Support Vector Classifier, and a deep learning baseline via Multi-Layer Perceptron (MLP) were rigorously tested using the same datasets.
Hyperparameter Optimization
Multiple optimization techniques were applied to fine-tune the parameters of the various models:
- Random Forest and Decision Tree: Parameters such as the number of estimators and tree depth were fine-tuned.
- K Nearest Neighbors: Enhanced through neighbor counts and distance weighting.
- Support Vector Classifier: Evaluated using both linear and RBF kernels.
- MLP Configuration: Involved testing varied hidden layer sizes and regularization strengths.
Final performance metrics from this comprehensive evaluation were derived through stratified ten-fold cross-validation, ensuring each model’s stability and generalizability were aptly assessed.
Through these detailed datasets and methodologies, RareNet stands as a significant advance in the realm of cancer research, with the potential to contribute meaningfully to rare cancer diagnostics and treatment strategies.