Unveiling the Datasets Behind Glioma Research
In the realm of cancer research, the quality and breadth of datasets are paramount. Among the leading sources of clinically relevant data is The Cancer Genome Atlas (TCGA), a rich repository hosting molecularly characterized tumors from over 11,000 patients across 33 different types of cancer, including multiple subtypes of glioma. This substantial body of work enables researchers to delve deep into the genetic underpinnings of various malignancies, driving forward the quest for improved treatment protocols.
The Cornerstone: TCGA and Glioma
To create a comprehensive dataset for analyzing primary diffuse gliomas, researchers have harnessed clinical data, somatic mutations, and gene expression profiles. For instance, the LGG and Glioblastoma Multiforme (GBM) datasets from TCGA are pivotal. These datasets have been effectively merged, enabling a consolidated analysis termed GBMLGG. Data is accessible through platforms like cBioPortal and the University of California, Santa Cruz Xena browser, which facilitate user-friendly exploration of this extensive genomic information.
In addition to the core TCGA data, researchers often enrich it by incorporating supplementary information from other projects. In this context, clinical data gaps within the GBMLGG dataset have been filled by utilizing datasets from TCGA’s lower-grade glioma (LGG) and glioblastoma (GBM) datasets. Such integration helps mitigate issues related to missing clinical data, ensuring a more robust dataset for analysis.
Including Longitudinal Insights: The GLASS Consortium
As an independent validation source, data from the Glioma Longitudinal Analysis (GLASS) Consortium plays a critical role. This collaborative initiative is focused on collecting and analyzing longitudinal genomic data from glioma patients. By designing protocols that incorporate surgical timelines, GLASS eliminates the complexities associated with disease progression filtering that often plague static datasets like TCGA.
The GLASS dataset contributes value through its more granular tracking of tumor recurrence, allowing for precise definitions of time to recurrence (TTR). This metric—which is calculated as the elapsed days between the surgery for a primary tumor and the first surgery for its recurrence—enhances our understanding of the dynamics of tumor progression in glioma patients.
Refining Dataset Quality: Inclusion Criteria and Histological Adjustments
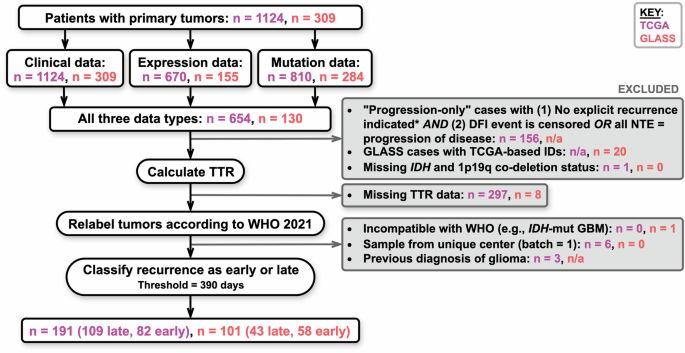
To assure only relevant data is analyzed, stringent inclusion criteria were set for both TCGA and GLASS datasets. For TCGA patients, filtering was executed to include only those with explicit progression indicators and the absence of a disease-free interval, ensuring that the final dataset authentically represents the progression of glioma.
Additionally, the classification of tumors has evolved alongside advancements in our understanding of glioma biology. The introduction of the 2021 WHO Classification of Tumors of the Central Nervous System necessitated updates to tumor labels based on mutations and deletions, such as IDH mutations and 1p19q codeletion statuses. A systematic approach was undertaken to relabel tumors according to these new guidelines, ensuring that the data maintained relevance to contemporary clinical practice.
Data Preprocessing: A Crucial Step
Prior to analysis, the gene expression data underwent log2 transformation and mean normalization to eliminate variability that might arise from different data acquisition methodologies. Furthermore, mutation data was scrutinized to calculate the incidence of non-silent mutations per gene per patient, generating a comprehensive genomic feature set for each individual.
Notably, clinical features—from patient age to tumor type—were refined and encoded. This rigorous preprocessing not only enhances the integrity of the dataset but also allows for a more precise modeling of tumor behavior and treatment response.
Arrival at the Final Patient Cohort
The ultimate goal of these efforts culminated in a final analysis cohort. For TCGA, a total of 191 patients were identified as meeting all outlined criteria. Of those, a significant proportion exhibited varied recurrence timelines, with 42.9% having recurrences occurring within 390 days post-initial treatment. This stratification provides valuable insights into the disease’s natural history and implications on treatment strategies.
Similarly, the GLASS dataset contributed a cohort of 101 patients, with recurrence timelines heavily favoring early recurrences. Such stark contrasts in recurrence dynamics between the two datasets reflect the need for tailored approaches in glioma treatment, reinforcing the importance of robust data in guiding therapy.
Ensuring Reliability: Statistics and Reproducibility
To ensure the reliability of the training and testing cohorts, statistical analyses were performed to evaluate common clinical attributes across different datasets. Techniques such as Chi-square tests and Mann-Whitney U tests were employed to validate that distributions remained comparable, asserting the robustness of the dataset utilized for glioma recurrence classification.
LUNAR: The Next Frontier
Thus far, the groundwork laid by these extensive datasets has enabled the development of gLunar—the glioma recurrence attention-based classifier. Leveraging clinical, gene expression, and mutation data, LUNAR utilizes a sophisticated architecture to analyze and predict early versus late recurrences in glioma patients.
The model incorporates attention mechanisms to capture relationships within and across different modality types, ultimately enhancing predictive accuracy. This innovative approach underscores how advanced statistical and machine learning techniques can be effectively deployed to glean deeper insights from complex behavioral patterns observed in glioma patients.
In conclusion, the assembly and meticulous processing of datasets such as TCGA and GLASS enable substantial advancements in our understanding and treatment approaches to gliomas, paving the way forward with innovative models like LUNAR. Such integrative strategies hold the potential to transform the landscape of cancer care, offering hope for better prognostic assessments and tailored interventions.