Exploring Drug-Target Interaction Datasets: An In-Depth Look at DrugBank, Davis, and KIBA
The field of drug discovery relies heavily on datasets that capture drug-target interactions (DTIs). Comprehensive datasets are essential for training and validating models aimed at predicting the interactions between drugs and their protein targets. In this context, we examine three pivotal datasets: DrugBank, Davis, and KIBA. Each offers unique insights and serves distinct roles in the realm of medicinal chemistry and pharmacology.
DrugBank Dataset
The DrugBank dataset is a rich repository for the pharmacological research community. The data used for this evaluation was published on January 3, 2020, and is classified as version 5.1.5. It originally consisted of a wide array of compounds, but extensive filtering was conducted to ensure the quality and relevance of the data. Notably, inorganic compounds and very small molecule compounds with unrecognized SMILES (Simplified Molecular Input Line Entry System) strings were excluded. Furthermore, any drugs with SMILES lengths exceeding 300 characters were also removed to meet the input constraints of MG-BERT, a well-known pre-trained model for bioinformatics.
After this meticulous cleansing process, the dataset yielded 6,148 drugs and 4,085 targets, culminating in a total of 16,531 known drug-target interaction pairs. These pairs served as the foundation for generating training samples. To balance the dataset, random negative samples were instituted by swapping the targets of drug-target pairs while ensuring no overlap with existing pairs. Ultimately, this effort resulted in 33,062 pairs, establishing an even positive-to-negative sample ratio of 1:1.
Davis and KIBA Datasets
In contrast, the Davis and KIBA datasets are tailored for assessing the binding affinity of drugs to proteins, specifically offering wet-lab values that can elucidate interaction strength. The datasets implement thresholds (5.0 and 12.1) to delineate positive from negative samples, facilitating the construction of a binary classification dataset. This method allows for a clearer understanding of the nuanced relationships between various drug compounds and their target proteins.
These datasets are integral for fine-tuning models to predict drug-target interactions, especially when combined with the cold-start methodology. By selecting 10% of drugs from DrugBank and utilizing the associated DTI pairs as a test set, the resulting 3,170 cold-start test samples provided a valuable opportunity to evaluate model efficacy in real-world scenarios—a common challenge in drug discovery.
Unseen Data Prediction with FDA-Approved Drugs
In an innovative approach to assess out-of-domain performance, all new drugs approved by the FDA in 2022 were assembled from DrugBank. The collection included 37 new drugs—22 new molecular entities and 15 biologic applications—with an emphasis on molecular drugs while excluding any that appeared in the training set. This yielded 13 drugs, 22 targets, and 24 interaction pairs.
Similar to the method employed for DrugBank, negative samples were constructed to maintain parity, resulting in 48 pairs of samples for a robust assessment of the model’s predictive capability on unseen data. This out-of-domain testing serves as a critical measure of a model’s adaptability and utility in the ever-evolving landscape of pharmacology.
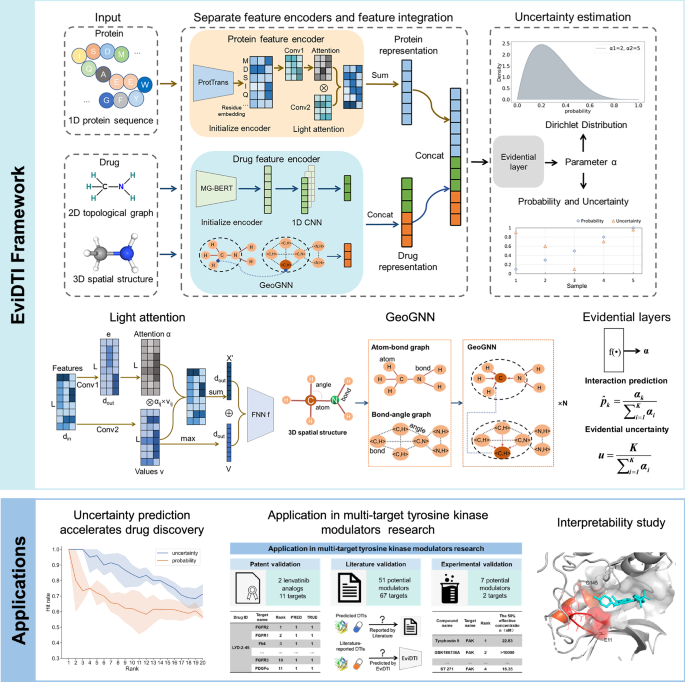
Framework Architecture: Understanding EviDTI
The framework architecture underpinning the analysis of these datasets is essential for effective prediction. The EviDTI model employs dual feature encoders for both proteins and drugs.
-
Protein Feature Encoder: Utilizing the ProtTrans model, the encoder extracts extensive sequence details, generating a feature matrix of size (L \times 1024), where (L) corresponds to the length of the protein chain. This information is processed through light attention mechanisms that enhance the focus on specific residues, thereby amplifying the model’s sensitivity to critical binding sites.
- Drug Feature Encoder: This encoder is bifurcated into two modules. The first handles 2D topological representations through the MG-BERT model, while the second delves into 3D structural complexities. By constructing atom-bond and bond-angle graphs, the model iteratively refines representation vectors, capturing intricate molecular dynamics and topological nuances vital for interaction predictions.
Evidential Learning: Moving Beyond Conventional Approaches
A salient feature of the EviDTI model is its innovative evidential learning approach. Unlike traditional models, which yield crisp probabilistic outputs, EviDTI operates using Dirichlet distributions to encapsulate uncertainty in predictions. Such a paradigm shift allows the model to provide not only a prediction but also an accompanying measure of confidence. This is particularly beneficial when evaluating novel drug interactions where data may be sparse, offering a layer of robustness that is vital for high-stakes decision-making in drug development.
The output of this model comprises belief masses, which quantify the support from the data for each interaction class, coupled with uncertainty levels that can inform researchers about the reliability of these predictions.
Comparative Analysis with Existing Models
To contextualize the performance of EviDTI, it is essential to consider various competing methods within the same landscape. Models such as DeepConv-DTI, GraphDTA, MolTrans, and several others have been employed to extract features from drugs and proteins through diverse architectures, ranging from convolutional networks to graph-based approaches. By retraining these models on the same datasets and performing meticulous hyperparameter tuning, a fair comparative baseline is established, allowing for an accurate assessment of EviDTI’s capabilities.
Statistical Analysis of Binding Sites
To validate the model’s predictions further, statistical analysis of binding sites is undertaken using real three-dimensional structures from the Protein Data Bank (PDB). The analytical rigor here involves correlating residues with higher attention values in the light attention (LA) mechanism to actual binding sites within the protein structure. Rigorous methodologies ensure only relevant structures and data are employed for this analysis, bolstering the credibility of the insights drawn.
Experimental Design and Validation
The EviDTI model, built upon a robust technical foundation, is implemented using Python and several prominent libraries. Rigorous experimental settings dictate the parameters for training, including batch size and optimizer configurations, ensuring reproducibility and reliability in performance evaluations.
Additional experiments, particularly in vitro kinase activity assays involving potential tyrosine kinase modulators, further substantiate the predictive accuracy of the model. These assays provide critical real-world insights into pharmacological interactions and help validate findings derived from computational predictions.
Implications for Future Research
The advancements in datasets like DrugBank, Davis, and KIBA, coupled with innovative modeling frameworks such as EviDTI, represent significant strides in drug discovery. The ability to predict drug-target interactions with enhanced accuracy and confidence signals a promising frontier in pharmacology and personalized medicine—ultimately paving the way for more effective therapeutics tailored to individual patient profiles. As research methodologies continue to evolve, the synergy between robust data analytics and machine learning promises unprecedented opportunities in the pharmacological domain.