Dataset Description and Analysis of COVID-19 Severity in Pediatric Patients
Overview of the Study
The present study embarked on a retrospective analysis of a hospital-based registry database focused on COVID-19, covering the period from February 25, 2020, to November 15, 2021. This analysis was conducted using data collected from the Children Hospital Medical Center, a specialized pediatric referral institution located in Tehran, Iran. Ethical approval was granted by the Ethics Committee of Tehran University of Medical Sciences (IR.TUMS.CHMC.REC1399.069). Informed consent was obtained from all participants and their legal guardians prior to data collection, ensuring adherence to the Declaration of Helsinki.
Dataset Composition
The registry database comprised 93 primary features, categorized into four main classes:
- Demographics: 3 features
- Clinical Characteristics: 40 features
- Comorbidity History: 6 features
- Laboratory Results: 43 features
An additional outcome variable was classified into severity status: 0 representing non-severe, and 1 indicating severe cases. Quantitative parameters were recorded numerically, while nominal parameters were treated as binary responses (1: Yes or 0: No). This comprehensive structure provides valuable insights into the patient profiles under study, as detailed in Supplementary Table 2.
Definition of Severe COVID-19
The study defined severe COVID-19 based on objective clinical outcomes, adapted from the World Health Organization (WHO) clinical progression scale. Severe disease was characterized by one or more of the following criteria:
- ICU admission for close monitoring
- Intubation due to respiratory failure
- Mechanical ventilation for support
- Death attributable to COVID-19 complications
These criteria align with prior peer-reviewed studies, thus ensuring the classification reflects clinically significant disease progression. The resulting severity status was utilized as the outcome variable (Y) in supervised learning models aimed at distinguishing between non-severe and severe cases.
Data Collection Process
Each patient was registered once in the dataset, with the first set of laboratory results collected during their initial admission. Demographic and comorbidity history was gathered from medical records or directly from patients and guardians, while clinical features were documented at the time of admission. Laboratory tests occurred during the initial hours of hospitalization, and chest CT imaging was performed at the discretion of a specialist, focusing on key findings such as ground-glass opacities and consolidations.
Chest CT Imaging
CT imaging served as an important diagnostic tool, with identifiable features including:
- Ground-glass opacities
- Consolidations
- Opacity distribution
- Pleural effusions
- Fibrosis and nodules
CT images were independently reviewed by two radiologists to ensure accuracy, with discrepancies resolved through consultation with a senior radiologist.
Data Pre-Processing
Prior to the application of machine learning algorithms, thorough data pre-processing was conducted to enhance data quality and reliability. Records with over 60% missing data were excluded. For remaining records, missing binary variable values were imputed using logistic regression, while continuous variables were addressed via predictive mean matching using the ‘mice’ package in R.
All continuous variables underwent Z-score normalization to promote consistency across features. Various categorical variables, such as blood group, were transformed into dummy variables, enhancing their suitability for machine learning models.
Inclusion and Exclusion Criteria
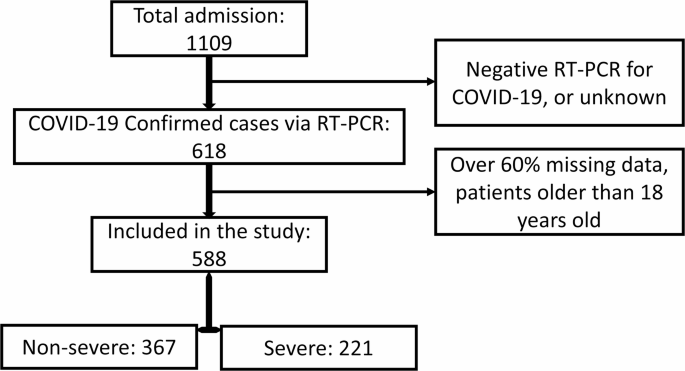
Inclusion in the study was restricted to children with positive RT-PCR COVID-19 test results. Exclusion criteria included:
- Negative RT-PCR test results
- Unknown patient dispositions
- Over 60% missing data
- Patients older than 18 years
A visual representation of the patient selection process can be found in Fig. 1, illustrating how 588 pediatric patients were included for subsequent predictive modeling analyses.
Feature Selection and Model Training
To mitigate overfitting in machine learning algorithms, the selection of key variables closely associated with the outcome variable was imperative. This study explored important predictors of COVID-19 severity through Generalized Boosted Models (GBM) and Random Forest (RF) algorithms. Highly correlated variables were removed to refine the feature set, ensuring robust predictive models.
Machine Learning Algorithms Employed
A diverse set of machine learning algorithms was employed, including:
- Neural Networks (NN)
- Generalized Boosted Models (GBM)
- Random Forest (RF)
- Recursive Partitioning and Regression Trees (RPART)
- k-Nearest Neighbors (k-NN)
- Kernel Support Vector Machines (KSVM)
Each algorithm was selected based on its strengths in medical predictive modeling, with NN chosen for its capacity to handle complex relationships, and GBM and RF for their robustness in high-dimensional data.
Performance Evaluation
The models used a binary classification approach, with performance metrics evaluated based on accuracy, sensitivity, and specificity. The dataset was split into a training set (70%) and a holdout set (30%) for evaluation.
Through the “SuperLearner” framework, individual models were combined using weighted averaging to leverage their strengths effectively. This approach, alongside internal cross-validation, enhanced performance estimation and generalization capabilities.
Hyperparameter Tuning and Class Imbalance Considerations
Hyperparameter tuning was generally not employed, with default parameters used for most models. Notably, the study addressed moderate class imbalance without re-sampling techniques or class weighting adjustments, relying instead on the SuperLearner’s capabilities to maintain balanced performance metrics across models.
Methodological Insights
To enrich understanding of the machine learning models’ functionality, a brief overview of their mathematical formulations is provided:
- Random Forest (RF): An ensemble of decision trees predicting the mode of individual tree outcomes.
- Gradient Boosted Machines (GBM): Trees built sequentially, minimizing residual errors.
- Support Vector Machines (SVM): Algorithms aiming to find optimal hyperplanes for class separation.
- Neural Networks (NN): Models utilizing layers of weighted connections to compute output.
Software and Tools
Data analysis and model training were conducted utilizing R version 4.4. The robust statistical computing environment ensured reproducibility and reliability in the results produced.
In summary, the detailed methodology presented here offers an in-depth exploration of the dataset and analytical approaches used to investigate COVID-19 severity among pediatric patients in Tehran. The findings have significant implications for understanding the clinical characteristics and outcomes associated with this virus in children, serving as a foundation for further research and clinical applications.