“Machine Learning and Image Analysis for Automatic Detection of Sister Chromatid Exchanges”
Machine Learning and Image Analysis for Automatic Detection of Sister Chromatid Exchanges
Understanding Sister Chromatid Exchanges (SCEs)
Sister chromatid exchanges (SCEs) are genetic processes where identical sister chromatids swap segments of DNA. This mechanism plays a critical role in genetic diversity and repair processes, making its detection vital in cell biology and genetics. For instance, in cancer research, understanding SCEs can help assess DNA damage and repair pathways.
Importance of Automatic Detection
Automatic detection of SCEs is essential due to the complexity and volume of data generated from chromosome analysis. Manual assessment is labor-intensive and prone to errors, leading to inconsistent results. Machine learning and image analysis can significantly enhance accuracy and efficiency, allowing for high-throughput screening in clinical and research settings.
Core Components of the Detection Process
The detection process can be broken down into several core components: image acquisition, data preprocessing, segmentation, and classification. Image acquisition involves capturing high-resolution images of chromosomes, often using advanced microscopy techniques. Data preprocessing cleans and normalizes these images to ensure accurate analysis. Segmentation isolates the chromosomes from the background, while classification assigns labels based on the presence of SCEs.
Step-by-Step Detection Process
-
Image Acquisition: High-resolution images of chromosomes are captured using fluorescent microscopy. For example, TK6 cells treated with specific dyes can reveal SCEs under ultraviolet light.
-
Preprocessing: Raw images undergo filtering to reduce noise and enhance contrast. Morphological operations may also be applied to sharpen edges and delineate chromosomes, ensuring they are accurately represented for further analysis.
-
Segmentation: Algorithms like Mask R-CNN are employed to identify and segment individual chromosomes from the processed images. This method utilizes deep learning techniques to improve the precision of chromosome delineation.
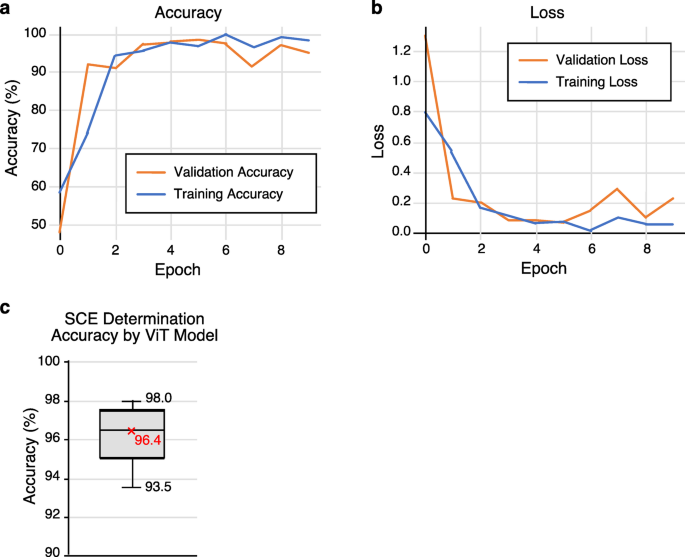
- Classification: The segmented chromosomes are analyzed using models such as ViT (Vision Transformer) to classify them based on the presence or absence of SCEs. The classification phase predicts SCE occurrences by analyzing features derived from the images.
Practical Scenario: SCE Analysis in Cancer Research
In cancer research, automatic SCE detection can yield insights into tumor characteristics and treatment efficacy. For instance, a study involving patient-derived tumor samples analyzed SCEs to assess chromosomal instability, a hallmark of cancer. By automating this evaluation, researchers could process thousands of samples with higher accuracy, accelerating the discovery of therapeutic targets.
Common Mistakes in Chromatid Analysis
A frequent error in SCE detection is inadequate segmentation, which can lead to misclassification of chromosomes. If overlapping or touching chromatids are not accurately separated, the results can skew data interpretation. To mitigate this risk, employing robust algorithms like Mask R-CNN with fine-tuning for specific datasets can vastly improve segmentation outcomes.
Tools and Metrics for Evaluation
The tools used for SCE detection include advanced imaging software and machine learning frameworks. Metrics such as Average Precision (AP) and the Analyzable Ratio (AR) are commonly applied to evaluate model performance. For example, a detection threshold might be set at a Confidence Score of 0.6 to balance sensitivity and specificity.
Alternatives in SCE Detection
Alternative methods involve using traditional staining techniques followed by manual analysis, which, though accurate, lack the throughput needed for large-scale studies. While machine learning methods offer higher speed and consistency, they may require extensive training datasets to realize optimal performance.
Frequently Asked Questions
Q: How does machine learning improve SCE detection?
Machine learning enhances SCE detection by automating the image analysis process, reducing human error, and increasing processing speed. Models can learn to recognize subtle differences between normal and exchanged chromatids based on extensive training.
Q: What are the limitations of using machine learning for SCE detection?
Limitations include the necessity for high-quality annotated training data and the potential for overfitting if models are not properly validated. It’s crucial to continually evaluate and refine models using diverse datasets.
Q: Can SCE detection be performed on other cell types?
Yes, SCE detection can be applied to various cell types, including normal somatic cells and tumor cells. Each application may require specific adjustments to the image analysis pipeline to account for differences in chromosomal structure.
Q: What advancements are being made in SCE analysis technology?
Current advancements include the integration of deep learning with emerging imaging technologies such as super-resolution microscopy, which allows for even greater detail and clarity in chromatid visualization, enhancing the accuracy of SCE detection.