Understanding the Datasets in Iron Deficiency Research
Introduction to Datasets
In the realm of medical research, the integrity and relevance of datasets are paramount. For an insightful study focusing on iron deficiency anemia (IDA) in Ghana, researchers gathered two distinct datasets. These datasets, integral to understanding demographic and health factors related to prospective blood donors and nulliparous women, provide a foundation for developing predictive models in personalized medicine.
Characteristics of the First Dataset
The first dataset was collected from prospective blood donors across three geographic regions in Ghana—Northern, Eastern, and Central. Notably, this dataset aimed to measure baseline ferritin levels among these individuals. While the Northern and Central regions offered rich, detailed data including variables like Age, Gender, WBC, HB, MCV, MCH, MCHC, PLT, HCT, CRP, and ferritin, the Eastern region data was less comprehensive, documenting only HB, MCV, and MCH. This inconsistency led to the exclusion of Eastern region data from further analysis.
The selection of specific variables was not arbitrary; metrics like MCV, MCH, MCHC, HB, and RBC have demonstrated utility in previous artificial neural network models aimed at detecting IDA. Although the original sample size was 190, preprocessing and exclusion of instances with CRP levels exceeding 5 mg/l—an indicator of inflammation—reduced the working sample to 188.
Insights from the Second Dataset
The second dataset shifts focus to preconception health, targeting nulliparous women aged 16 to 36 years across two zonal divisions in Ghana. With 336 instances, this dataset aimed to identify iron stores within this demographic, crucial for understanding ID prevalence before pregnancy.
Variables recorded included AGE, BMI, WHR, HB, MCV, MCH, MCHC, CRP, and serum ferritin (SF). The criteria to define iron deficiency was set at SF < 15µg/l. Within this dataset, 109 cases were identified as ID positive, while 207 were considered ID negative, reflecting patterns critical to understanding the broader challenges of iron deficiency in Ghana.
Ethical Considerations
All research protocols received ethical clearance from the University of Cape Coast, ensuring alignment with global ethical standards, including participant confidentiality and informed consent. Participants were fully briefed on their rights, encapsulating a commitment to their autonomy throughout the research process.
Introducing BamClassifier: Innovating ID Status Predictions
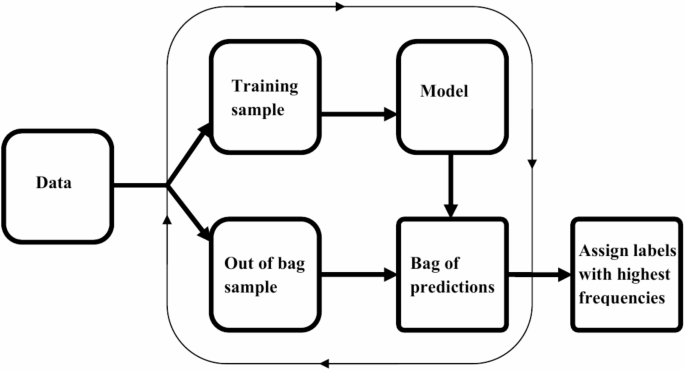
Machine learning techniques now play a pivotal role in precision medicine, particularly in ID assessments where standard methodologies may fall short. Introducing the BamClassifier, an innovative approach leveraging ensemble methods to reduce learning variance through bagging, effectively amplifies the process of identifying ID statuses.
Sub-sampling Strategies in Model Training
The BamClassifier methodology employs a systematic sub-sampling approach, randomly extracting equally sized samples without replacement from the original dataset. Each unique subsample generates a naive Bayes model, later tested on out-of-bag instances. By ensuring that every instance has the opportunity to contribute to both model building and validation, this technique enhances model reliability and robustness.
Constructing the BamClassifier: Step-by-Step
The core process involves multiple steps for constructing the BamClassifier:
- Establish the subsample size for model training.
- Randomly select a subsample from the original dataset.
- Develop a median-supplement naive Bayes model utilizing this subsample.
- Implement predictions on out-of-bag samples.
- Repeat this sampling and modeling procedure until every instance has been utilized.
- Aggregate predictions into a consensus bag for final classification.
This systematic approach not only balances the discrepancies between ID positive and negative instances but also fortifies the models against the complexities of biological data.
Naive Bayes Application in ID Diagnosis
Utilizing Bayes’ theorem, the probability calculations involved provide a robust framework for classifying ID statuses. The model assesses the probability of ID classification based on the features, integrating conditional independence assumptions that align with empirical data attributes.
Performance Metrics for Evaluation
To comprehensively gauge the effectiveness of the BamClassifier, performance metrics such as accuracy, specificity, and sensitivity serve as benchmarks when contrasted against other established algorithms, including logistic regression and random forests. The accuracy ratio reflects the proportion of correct classifications, while specificity and sensitivity shed light on the model’s strengths in identifying various ID statuses.
Statistical Analysis and Data Robustness
Rigorous statistical methods, such as the Shapiro test and Mann-Whitney test, were employed to assess data distribution and mean comparisons, ensuring the analysis adhered to scientific rigor. For comprehensive validation, a robust assessment of the BamClassifier’s performance was conducted through bootstrap sampling and various tests, which confirmed its efficacy across different scenarios.
Accessibility and Transparency
All code supporting the study’s analytical framework is available as supplementary material, underlining a commitment to transparency and reproducibility in the research process.
Conclusion
The deliberate design and comprehensive assessment of datasets illuminate the significant relationship between measurement, analysis, and health outcomes in the context of iron deficiency. By leveraging innovative methods like the BamClassifier, the research paves the way for enhanced predictive capabilities, potentially transforming how ID is diagnosed and managed in vulnerable populations.