“Improving Peptide Identification in Metaproteomics with Deep Learning and Curriculum Learning”
Improving Peptide Identification in Metaproteomics with Deep Learning and Curriculum Learning
Understanding Metaproteomics and Its Importance
Metaproteomics is the study of proteins in a community of microorganisms, providing insights into the functions and interactions within complex ecosystems such as human gut microbiomes. By analyzing protein profiles, researchers can determine how these communities respond to changes in the environment or health conditions, making metaproteomics crucial for fields like microbiome research, disease studies, and biotechnology.
For instance, metaproteomics has revealed how gut bacteria influence human metabolism and immune responses, leading to new avenues for treating conditions like obesity and diabetes. As the quest for understanding complex microbial interactions deepens, efficient and accurate peptide identification becomes increasingly important.
Key Elements of Peptide Identification in Metaproteomics
Peptide identification involves matching peptide sequences derived from mass spectrometry data against databases of known proteins. The core components include:
- Mass Spectrometry (MS): This technique analyzes the mass-to-charge ratio of ionized peptides to identify their sequences.
- Database Search Engines: Tools like Comet and Myrimatch are used to match detected peptides with sequences in databases. Each engine has unique algorithms and databases, impacting the identification outcome significantly.
- Statistical Validation: False discovery rates (FDR) are calculated to estimate the proportion of false identifications, ensuring reliability in results. Techniques such as the target-decoy strategy are often used.
Through these elements, researchers aim to identify proteins with high confidence, which can impact biological interpretations and follow-up studies.
Step-by-Step Process of Peptide Identification
Identifying peptides in metaproteomics typically follows a structured lifecycle:
- Sample Preparation: Microbial communities are extracted from samples (e.g., feces, soil) and digested into peptides using enzymes like trypsin.
- Mass Spectrometry Analysis: The resulting peptides are analyzed in a mass spectrometer, generating spectra for interpretation.
- Database Searching: Mass spectra are compared against protein databases using search engines. Each candidate peptide gets a score based on its match quality.
- Statistical Filtering: Identifications are statistically evaluated, commonly using FDR to control for false positives.
- Validation: Peptides are further validated against known literature or additional experiments.
This lifecycle ensures a systematic approach to peptide identification, but it can be prone to errors, particularly in complex samples.
Challenges and Solutions in Peptide Identification
Common pitfalls in metaproteomics include high false discovery rates, particularly in diverse microbial communities where incomplete databases may lead to overlapping peptide identifications. For example, a database that misses several key species would generate many false positives, distorting the biological interpretation.
To mitigate these issues, researchers are employing advanced methodologies such as deep learning through frameworks like WinnowNet. This model leverages machine learning to learn from existing data, helping to enhance the accuracy of peptide identification without traditional training processes.
Tools and Frameworks for Enhanced Identification
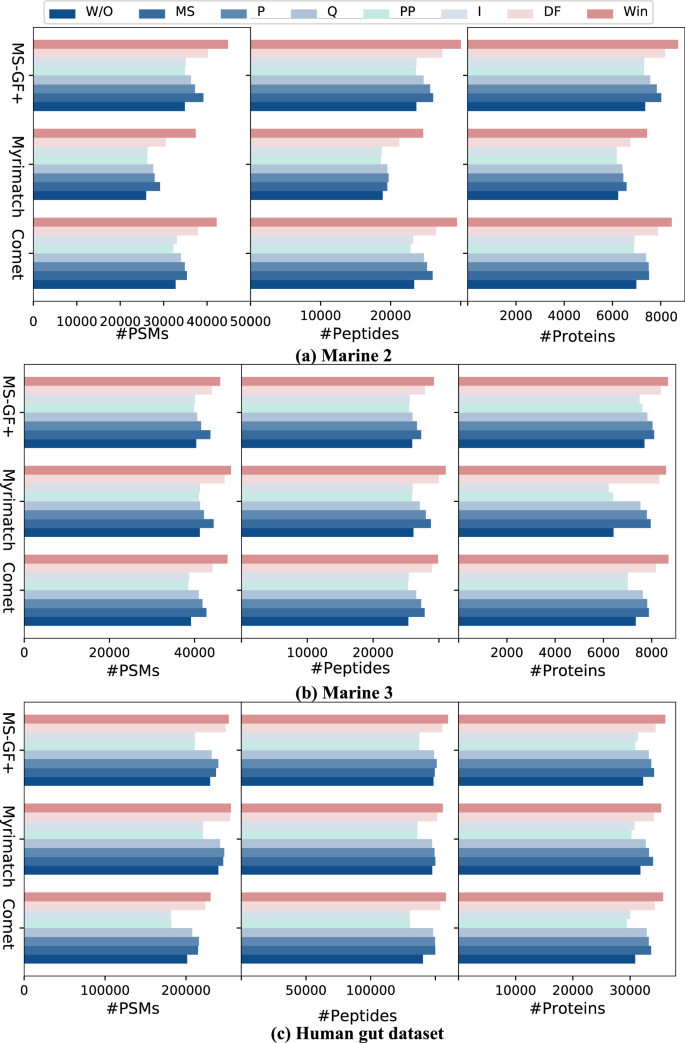
WinnowNet is a state-of-the-art framework that integrates deep learning into the peptide identification pipeline. It effectively combines features from peptide spectrum matches (PSMs) leveraging both convolutional neural networks and self-attention mechanisms. This hybrid approach automatically extracts discriminative features, significantly enhancing identification rates.
For instance, in recent evaluations, WinnowNet consistently outperformed traditional filtering methods across multiple datasets, identifying more peptides with lower false discovery rates. The model has shown improvements, with some experiments reporting up to 12% better identification rates compared to older systems like MS²Rescore.
Practical Application: Case Studies in Metaproteomics
Recent studies have utilized WinnowNet in various contexts, particularly focusing on different ecosystems, such as marine environments and the human gut. In a detailed analysis of gut microbiomes, WinnowNet identified numerous species previously overlooked, shedding light on their roles in human health.
These findings are crucial as they help in identifying specific microbial function pathways, ultimately contributing to our understanding of the microbiome’s impact on host health. For example, improved peptide identification has led to insights on how specific microbes contribute to immune modulation and metabolic processes.
Trade-offs and Alternatives in Peptide Identification
While deep learning models like WinnowNet offer enhanced identification capabilities, they also come with trade-offs. The computational requirements for training such models can be significant, necessitating advanced hardware and expertise. In scenarios where resources are limited, traditional techniques such as Percolator may still be preferable.
Conversely, using WinnowNet offers the potential for higher identification accuracy and lower false rates, thus it is advisable for researchers with access to adequate computational resources. The decision between methods should be tailored to the specific research goals and available infrastructure.
Frequently Asked Questions
What is the significance of false discovery rates in metaproteomics?
False discovery rates are crucial as they help determine the reliability of peptide identifications. A controlled FDR ensures that researchers can trust their findings, particularly in complex samples where misidentification can lead to erroneous biological conclusions.
How does curriculum learning enhance deep learning in this context?
Curriculum learning allows models to learn from simpler to more complex data, improving their ability to generalize across varied datasets. This strategy has shown to enhance the robustness of peptide identification, particularly in diverse metaproteomic studies.
What role do database search engines play in peptide identification?
Database search engines act as the primary tool for matching detected peptides against known sequences, forming the foundation of the identification process. The accuracy of these engines directly impacts the reliability of the results.
What is the trade-off of using deep learning models like WinnowNet?
While models like WinnowNet provide improved accuracy, they require significant computational resources and expertise in machine learning. Researchers should weigh these factors against their project’s specific needs and capacities.