Identifying Immune Infiltration Subtypes via Consensus Clustering
In the quest to understand and treat breast cancer effectively, researchers have turned their attention to the immune landscape of tumors. A recent study harnessed the power of consensus clustering to identify distinct immune infiltration subtypes within the TCGA-BRCA cohort. This technique not only helps classify patients but also opens avenues for targeted therapies based on immune responses.
Calculation of Enrichment Scores
To kick things off, the researchers calculated enrichment scores for 28 different immune cell types within the TCGA-BRCA cohort using a method known as single-sample Gene Set Enrichment Analysis (ssGSEA). This allowed them to understand the immune composition present in individual tumors better.
Unsupervised Consensus Clustering
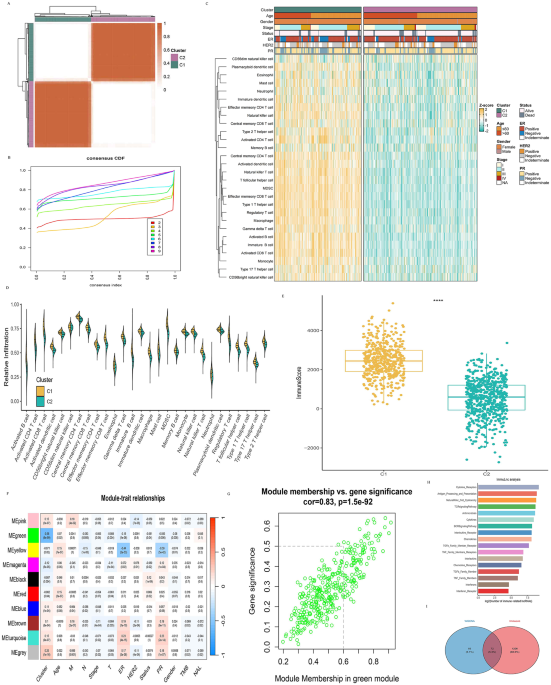
Following the scoring, an unsupervised consensus clustering approach was employed. Utilizing the area under the cumulative distribution function (CDF) curve, the researchers generated consensus heatmaps to ascertain the optimal number of clusters. When the number of clusters (k) was set to 2, a striking division became evident. Breast cancer patients were distinctly separated into two clusters, characterized by unique expression patterns.
The results were compelling: Cluster 1 (C1), which showed a significantly higher level of immune cell infiltration than Cluster 2 (C2), was defined as an immunologically rich tumor. Conversely, C2 was characterized as an immunologically poor tumor, highlighting the profound differences in immune activity within these subtypes.
To verify these observations, the researchers employed six different algorithms, including TIMER and quanTIseq, which consistently confirmed that C1 exhibited greater immune cell abundance than C2. Additionally, the ESTIMATE immune score further validated these findings, with C1 yielding higher scores than C2.
Construction of WGCNA and Identification of Immune-Related lncRNA Modules
Understanding how immune response correlates with gene expression is another critical aspect of breast cancer research. To delve deeper, the researchers constructed a weighted gene co-expression network analysis (WGCNA), setting parameters for a scale-free network to 0.55. Utilizing this methodology, they identified ten gene expression modules that demonstrated varying correlations with clinical traits, such as immune clusters, tumor characteristics, and patient demographics.
Among these modules, one stood out for its robust correlation with clinical characteristics: the Immune Cluster module. The correlation coefficient reached an impressive 0.96, affirming the reliability of lncRNA module construction. The lncRNAs within this key module became the focus for developing models centered on immune-related factors.
ImmuLncRNA Algorithm Screening for Immunity-Related lncRNAs
The researchers then leveraged the ImmuLncRNA algorithm to extract lncRNA regulatory factors involved in immune regulation. This function aided in unveiling 1,278 unique immune-related lncRNAs. Most of these were found to play roles in critical immune pathways, including TCR signaling, cytokine receptors, and natural killer cell cytotoxicity.
To refine their results further, they intersected the lncRNAs identified by the ImmuLncRNA algorithm with those discovered through WGCNA, culminating in a definitive list of 72 immune-related lncRNAs.
Construction of a Prognostic Immune-Related lncRNA Signature
A univariate Cox regression analysis on these 72 lncRNAs revealed 15 significant prognostic factors for breast cancer patients. The researchers, utilizing machine learning algorithms, constructed a prognostic model, achieving an optimal C-Index of 0.71 with features selected through Lasso regression and StepCox.
From this analysis, 9 lncRNAs emerged as crucial prognostic biomarkers. Each patient’s risk score was meticulously constructed using the expression levels of these lncRNAs multiplied by their respective coefficients derived from regression analyses.
Evaluation of the IRLS Model
The team tested the predictive performance of their newly established Immune-Related lncRNA Score (IRLS) model using various statistical indicators across multiple datasets. Impressively, the AUC values for predicting one-, three-, and five-year overall survival (OS) were consistently high across different cohorts, affirming the reliability of this model.
Notably, this model not only showcased promising results in the TCGA-BRCA dataset but also validated its effectiveness in other datasets such as GSE58812 and GSE103091, signifying robust predictive power.
Comparative Assessment with Established Models
In a comparative analysis of the IRLS with 95 other gene expression prediction models, IRLS uniquely demonstrated significant differences in predicting overall survival across eight datasets. The IRLS model stood out due to its consistent performance, unlike many other models that faltered outside their training sets.
Predictive Value for Treatment Regimens
When exploring the utility of the IRLS score in treatment responses, the researchers included datasets associated with various therapies. Their investigations revealed that patients responding to treatments, such as Paclitaxel and the AFG combination regimen, exhibited significantly different IRLS scores, indicating potential as a biomarker for treatment efficacy.
Implications for Immunotherapy
Further research highlighted the correlation between IRLS scores and immune cell infiltration, a key factor in determining the success of immune checkpoint inhibitors (ICIs). Low IRLS scores were consistently associated with higher levels of immune cell markers like CD8A and PD-L1. Notably, lower IRLS scores appeared predictive of greater immune activity, underscoring the model’s relevance in the context of ICI therapy.
Conclusion on the Future of IRLS
The advancements in identifying immune infiltration subtypes and constructing a prognostic model have significant implications for personalized medicine in breast cancer. By integrating immune profiling with lncRNA analysis, this pioneering study paves the way for future research aimed at optimizing treatment strategies and improving patient outcomes.