Dataset Description: A Deep Dive into Brain Tumor Segmentation and Classification
Introduction to Datasets
In medical imaging, the ability to accurately segment and classify brain tumors is crucial for effective treatment planning. This article focuses on two prominent datasets utilized for such tasks: the BraTS2020 (Multimodal Brain Tumor Segmentation) dataset and the Figshare brain tumor dataset. Both datasets offer invaluable resources for researchers and practitioners aiming to improve diagnostic accuracy and patient outcomes.
BraTS2020 Dataset
The BraTS2020 dataset is a comprehensive medical imaging repository that’s widely regarded as the gold standard for brain tumor segmentation tasks. This dataset primarily focuses on gliomas, which are recognized as the most common type of brain tumor.
Structure and Features
The BraTS2020 dataset consists of pre-operative MRI scans that present a variety of imaging sequences, specifically:
- T1-weighted images
- T2-weighted images
- T1-weighted with contrast enhancement (T1-CE)
- Fluid-attenuated inversion recovery (FLAIR)
A critical aspect of this dataset is its expert annotations, which clearly delineate tumor components such as:
- Necrotic core
- Enhancing tumor
- Peritumoral edema
In total, the dataset includes 369 labeled MRI cases divided into:
- 80% for training
- 10% for validation
- 10% for testing
This structured division ensures robust training and evaluation of machine learning models, aiding in the precise segmentation and classification of gliomas, as highlighted in Table 9.
Figshare Brain Tumor Dataset
The Figshare brain tumor dataset serves as a complementary resource. With a collection of 3064 T1-weighted contrast-enhanced MRI scans, this dataset encompasses images from 233 individuals, featuring three distinct types of brain tumors:
- Glioma slices (1426)
- Meningioma slices (708)
- Pituitary tumor slices (930)
The Figshare dataset’s accessibility makes it an appealing choice for researchers focusing on brain tumor classification, further divided similarly into training, validation, and testing datasets.
Data Processing Techniques
Both datasets focus on preprocessing to ensure efficacy in model training.
Image Loading and Transformation
To prepare the BraTS images for model input, the NiBabel Python library is utilized, which handles various neuroimaging file formats. These images are converted into 2D arrays with the help of the Numpy library.
For model training, frameworks such as TensorFlow, Keras, and Scikit-learn are employed. The images are resized to a uniform 256×256 pixels, making them compatible with the proposed Hybrid Vision U-Net (HVU) architectures. Data augmentation techniques—rotation, scaling, and flipping—are also implemented to enrich the dataset with diverse samples, further enhancing model training.
Hybrid Vision U-Net Architecture (HVU)
The Hybrid Vision U-Net (HVU) architecture serves as a pioneering framework in deep learning for brain tumor segmentation and classification. This architecture uniquely integrates the strengths of pre-trained Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs), optimizing both local feature extraction and global context modeling.
Architecture Variants
The HVU architecture features four variants, each named according to the pre-trained CNN integrated into the model:
- ResVU-ED
- VggVU-ED
- XceptionVU-ED
- DenseVU-ED
These variants employ distinct pre-trained models (ResNet50, VGG16, Xception, and DenseNet121) to maximize segmentation capabilities, ensuring the framework is adaptable to various imaging tasks.
Features of the HVU Architecture
ResVU-ED
This variant fuses ResNet with ViT and U-Net, leveraging the strength of residual blocks for capturing contextual data vital for pixel-wise segmentation. It utilizes the first 16 layers of ResNet50, offering a total of 4,17,43,876 parameters.
VggVU-ED
By coupling the VGG16 model with ViT and U-Net, VggVU-ED provides robust feature extraction. The features taken from its first 17 layers emphasize segmentation efficacy, accommodating 5,21,34,596 parameters.
XceptionVU-ED
Integrating Xception layers within the U-Net architecture, this model benefits from depthwise separable convolutions to capture detailed patterns, totaling 4,85,14,732 parameters.
DenseVU-ED
This variant employs DenseNet principles to promote efficient learning through dense connectivity, using the first 5 layers for feature extraction, which accounts for 3,69,57,380 parameters.
Vision Transformer Integration
The architecture of Vision Transformers (ViT) enhances global representation capabilities through self-attention mechanisms. By dividing input images into fixed-size patches, ViT learns relationships between different regions, facilitating tumor identification across diverse shapes and locations.
Self-Attention Mechanism
In ViT, each image patch is converted into a flattened 1D vector and projected into a lower-dimensional embedding space. Positional embeddings maintain spatial context, enabling the model to discern patch positions effectively.
Feature Integration with U-Net
The U-Net structure acts as a backbone in the HVU framework. Its encoder compresses input images through a series of convolutional and max-pooling layers, while the decoder restores spatial resolution by upsampling compressed features.
Skip Connections
Skip connections between corresponding encoder and decoder layers help preserve fine-grained details critical for accurate localization of anatomical structures.
Fusing Features
At the bottleneck layer, features from the various pre-trained models and the Vision Transformer are merged, enhancing the overall representation and promoting accurate segmentation outputs.
HVU-ED Segmenter
The architecture of the HVU-ED segmenter is specifically designed for multi-class segmentation tasks, leveraging the strengths of the U-Net framework.
Model Specifications
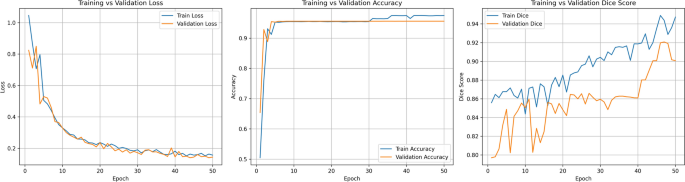
Preprocessed images sized 256×256 are employed as input. The segmentation results demonstrate the effectiveness of the model in delineating tumor boundaries, as illustrated in Fig. 12.
Training Protocol
Using the Adam optimizer with a learning rate of 0.001, the model boasts competitive performance metrics. Evaluation involves calculating the dice score, accuracy, and other standard metrics to validate segmentation effectiveness.
HVU-E Classifier
The HVU architecture identifies and segments tumors, but it can also be adapted for classification tasks. By modifying the decoder, the HVU-E classifier effectively categorizes images based on tumor type.
Classification Process
The U-Net encoder gathers important local features necessary for class distinction, while the bottleneck captures more complex features from the ViT. The combined features undergo flattening and are fed into dense layers for classification.
In summary, the BraTS2020 and Figshare datasets offer rich, nuanced resources for advancing brain tumor segmentation and classification. The innovative HVU architecture, with its hybrid approach merging CNN and ViT capabilities, demonstrates significant potential in this critical area of medical imaging. The structure not only enhances the accuracy of tumor segmentation but also sets a strong foundation for future advancements in medical AI.