Investigating Fluorescence Imaging and Deep Learning in Fungal Detection
Materials
In an innovative study conducted under the guidance of the Ethics Committee (KY2023-057) at Huashan Hospital, affiliated with Fudan University, researchers embarked on a mission to advance our understanding of superficial fungi through fluorescence imaging. The framework of this study adhered rigorously to the ethical principles outlined in the Declaration of Helsinki, ensuring that all examinations were conducted with the patients’ informed consent regarding their clinical data and sample processing. The collection of clinical samples was conducted methodically from February 2023 to February 2024, without requiring personal information from participants.
The fluorescence staining solution utilized for fungal imaging was supplied by Jiangsu Life Time Biological Co., Ltd. Image scanning was impressively performed at 10× magnification using a state-of-the-art intelligent fluorescence microscope from Shanghai TuLi Technology Co., Ltd., showcasing the integration of advanced technology in clinical research.
Annotation and Dataset
The collected images, each boasting a high resolution of 1,920 × 1,080 pixels, underwent meticulous annotation by two experienced clinicians using an in-house developed software tool named APTime, brought to life by SODA Data Technology Co., Ltd. A total of 942 images were labeled, forming the foundation for an object detection model. Among these images, 813 were strategically partitioned into training and validation sets, while 129 served to construct an independent test set, fostering a robust framework for model evaluation.
To fine-tune the mycelium detection model, 689 images featuring mycelium were gathered and meticulously cropped into 600 × 600-pixel tiles with a 160-pixel overlap. This resulted in a significant dataset comprising 1,351 mycelium-positive patches and 3,072 mycelium-negative patches. These datasets were systematically organized into training, validation, and testing sets, as detailed in the accompanying table.
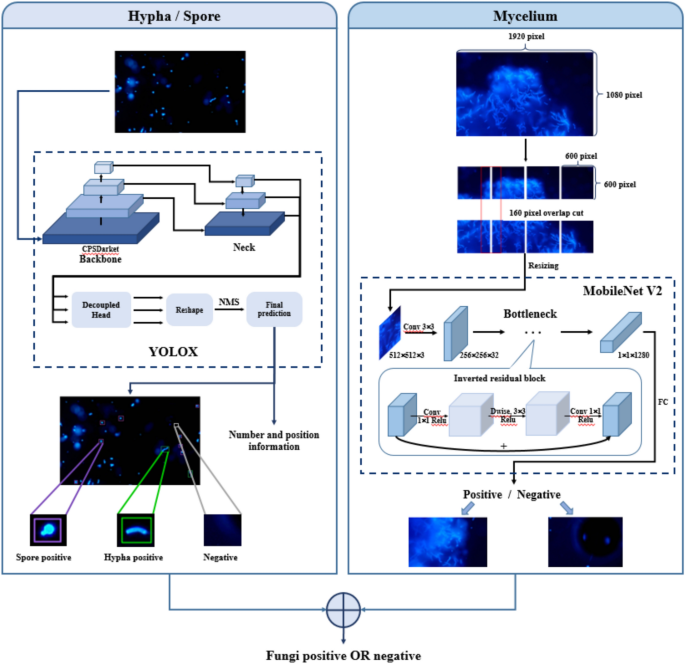
Dual-Model Framework
The innovation in this study is captured through a dual-model framework that leverages fluorescence fungal images analyzed by two deep learning models. This confluence of technologies allows for a comprehensive output derived from integrating the results of both models. The YOLOX model, specifically the YOLOX-L variant, is employed first to detect scattered hyphae and spores. It excels in classifying, locating, and counting fungal spores and hyphae, providing crucial insights into their distribution and prevalence.
For a more nuanced analysis of mycelium, the MobileNet V2 architecture is deployed. The images initiate at the original size of 1,920 × 1,080 pixels and are meticulously segmented into 160-pixel tiles, again utilizing a 600 × 600-pixel overlap. After resizing to 512 × 512 pixels, the mobile architecture processes these tiles. The classification result for each image tile is aggregated such that if any tile indicates a positive result, the entire image is deemed mycelium-positive, demonstrating the model’s robust sensitivity.
Model Training
The preprocessing of images plays a pivotal role in the training phase, encompassing normalization through pixel value division by 255, preservation of standard RGB channel ordering, and implementing brightness adjustments of ± 20% to cater to variable illumination conditions.
Training the YOLOX-L model involves sophisticated data augmentation techniques including MixUp and Mosaic strategies. Meanwhile, the MobileNet model benefits from techniques such as random flips, rotations, and blurring, alongside tweaks in hue and saturation. Notably, batch-level balancing is utilized in the MobileNet training to ensure an equal representation of positive and negative samples, effectively addressing potential class imbalances.
To enhance generalization and mitigate overfitting, both models incorporate dropout rates, label smoothing, and L2 weight regularization during training. Collectively, these strategies forge a pathway for improved performance when encountering unseen data without compromising diagnostic accuracy.
Evaluation
A comprehensive evaluation framework was established to assess the efficacy of the proposed model. Critical evaluation metrics focused on various performance aspects: the accuracy of the YOLOX model in detecting spores and hyphae, the consistency of clinical interpretations, the classification accuracy of the MobileNet V2 model in identifying mycelium, and the overall performance of the dual-model framework in ascertaining sample positivity.
For the YOLOX model, metrics such as Intersection over Union (IoU) were utilized to measure the overlap between predicted and actual bounding boxes. Other performance metrics included precision, recall, PR-curve, and F1-score, which were pivotal in quantifying the model’s detection effectiveness. To gauge the consistency of interpretations among clinicians, the F1-score served as a valuable index. For the MobileNet V2 model and the overarching dual-model framework, precision, recall, Kappa, and F1-score metrics were employed, supporting a rigorous evaluation of the models’ capabilities.