Evaluating Lesion-Location Relationship Mapping Tasks with LLM and BERT
Introduction
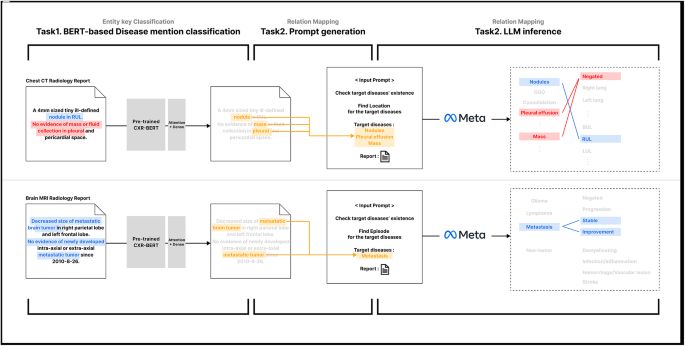
In the evolving landscape of medical imaging and reporting, there’s an increasing reliance on advanced natural language processing (NLP) techniques to extract meaningful insights from radiological reports. One significant exploration involves the performance evaluation of lesion-location relationship mapping tasks, specifically through a new approach utilizing large language models (LLMs) alongside BERT integration.
Performance Insights from Chest CT Scans
The evaluation of the lesion-location relationship mapping task was conducted at the lesion level using the BERT-LLM framework. The model produced an overall macro average accuracy of 56.13%, paired with an F1-score of 77.39. It’s crucial to note that while F1-score focuses on the ratio of correctly identified location matches within each lesion, the accuracy metric takes a stricter stance—where a match is only counted as correct if all locations are precisely identified for a particular lesion.
Interestingly, the model achieved its highest performance for mass lesions in chest CT scans, boasting an impressive accuracy of 80.00 and an F1-score of 89.55. Conversely, fibrosis lesions showed the lowest performance metrics, with an accuracy of just 40.82 and an F1-score of 64.00.
Precision vs. Recall
A deeper dive into the precision and recall of various lesions reveals that the model often exhibited higher precision than recall. However, certain lesions such as bronchiectasis, bronchial wall thickening, and interstitial thickening presented an anomaly where recall scores outperformed precision by margins of 5.09, 1.13, and 19.75 points, respectively. Particularly noteworthy was interstitial thickening, which not only showed the largest disparity but also landed as the fourth-lowest in terms of F1-score at 77.78—indicating the complexity of accurately identifying this condition in the report.
Rule-Based Method Benchmarking
A significant aspect of the evaluation was the comparison of the LLM with BERT approach against traditional rule-based methods. The LLM approach demonstrated a marked superiority across most lesion types and evaluation metrics, revealing an approximate improvement of 34.42 points in the macro-averaged F1-score.
The rule-based method seemed particularly deficient for specific lesions such as bronchial wall thickening and interstitial thickening, which resulted in a performance of zero in key metrics. This stark comparison underscores the potential advantage of leveraging LLM capabilities over conventional methods.
Analyzing Malignant Tumor Diagnosis via Brain MRI Reports
Shifting the focus to brain MRI, the evaluation of the diagnosis-episode relationship mapping task was similarly conducted using the LLM with BERT approach. The model achieved an overall macro average accuracy of 63.12 and an F1-score of 70.58. Just like in chest CT evaluations, accuracy offered a stricter measure, whereas the F1-score depicted a more rounded understanding of the model’s capabilities.
In this context, the model excelled in episode matching for lymphoma diagnoses with an F1-score of 72.69, while malignant metastasis exhibited a lower F1-score at 69.07, showcasing a notable accuracy of just 48.5. This highlights some inherent complexity in categorizing tumors with multiple episode types, often leading to lower accuracy due to the fragmented nature of present data.
Precision and Recall Disparities
This evaluation also unveiled a concerning trend in precision and recall metrics. In the glioma diagnoses, a notable disparity emerged, where recall exceeded precision by 27.54 points, indicating a potential propensity for false positives. This suggests that the presence of diverse episode types within a single report might detract from overall precision, warranting a closer look at the multifaceted nature of these reports.
Patient Progression Assessment through Longitudinal Data Analysis
Another remarkable aspect of the research was assessing patient progression over time, deriving insights from a robust database comprising 7,096 patients and 27,028 reports. The data revealed an average of 3.8 reports per patient, with variances ranging from 1 to 43 reports.
The application of the two-stage pipeline for episode mapping confirmed an F1-score of 70.58, bolstered by a higher recall of 73.31, demonstrating the model’s efficacy in recognizing positive episode changes. Filtering out reports lacking malignant tumor diagnoses and excluding patients with fewer than three reports added rigor to the analysis.
Insights from Episode Changes
A detailed exploration of episode changes, as showcased in a Sankey diagram, illustrated how patients with metastasis and glioma predominantly experienced progression episodes first, in stark contrast to lymphoma patients, who commonly showed improvement episodes upfront. Across all diagnoses, there was a clear tendency for episodes to shift to different labels rather than remaining static, revealing insights into patient progression and treatment efficacy.
Enhancements via Entity Classification and Comparisons
The integrated pipeline presented a substantial macro-average F1-score improvement of 93.4 in entity classification tasks. In assessing performance, McNemar’s test confirmed that the integrated approach demonstrated statistically significant advancements over individual BERT and LLM models. This reinforces the value in crafting a framework where these two methodologies can complement each other effectively.
Analyzing Classification Behavior
A closer examination of individual models elaborated on their respective strengths and weaknesses. BERT exhibited remarkable sensitivity, generating only eight false negatives across various lesion types but presented with a concerning number of false positives, standing at 371 out of 3,960 predictions. This highlighted a critical trade-off, where the sensitivity in keyword detection comes at the cost of precision—a known challenge in BERT’s handling of negation in medical texts.
Bridging Semantic Contexts with Advanced NLP Techniques
The development of this two-stage pipeline transcends mere disease detection; it ventures into accurately mapping relationships between identified entities—be it lesions or episodes. The rule-based methods occasionally excelled in precision due to their stricter criteria for matching (i.e., requiring co-occurrence within the same sentence). However, this rigidity often resulted in missed relationships that spanned multiple sentences—an aspect where LLMs like those employed here thrive.
Existing Method Comparisons
Noteworthy comparisons to other research efforts highlight the competitive nature of this methodology. For instance, while CheXbert previously achieved a macro-average F1-score of 79.8, this study’s entity classification outstripped that figure with an impressive score of 93.4, reflecting the efficiency of the proposed approach even within constraints of limited supervision and smaller datasets.
Limitations and Future Directions
Despite the promise shown in this framework, there are noted limitations, particularly concerning model performance tethered primarily to the capabilities of the LLM utilized. Investing in larger-scale open-source models is anticipated to further maximize both entity classification and relationship matching efficiency.
Future endeavors should also focus on further validation across diverse reporting styles and integrating seamlessly with hospital systems for heightened clinical relevance. Exploring potential integration with various imaging modalities promises exciting avenues for advancement within this invaluable area of medical informatics.
Conclusion
By harnessing the strengths of LLMs alongside traditional models like BERT, this research lays the groundwork for a robust NLP pipeline tailored for radiology reports. The integration not only enhances accuracy in extraction tasks but provides meaningful insights into patient care, diagnosis, and treatment pathways. With ongoing advancements and iterations on this methodology, the aspiration remains to fortify the role of NLP in optimizing clinical practices.