Implementation Details of the Scalable Competitive SMLM Data Analysis Framework
Framework Compatibility and Overview
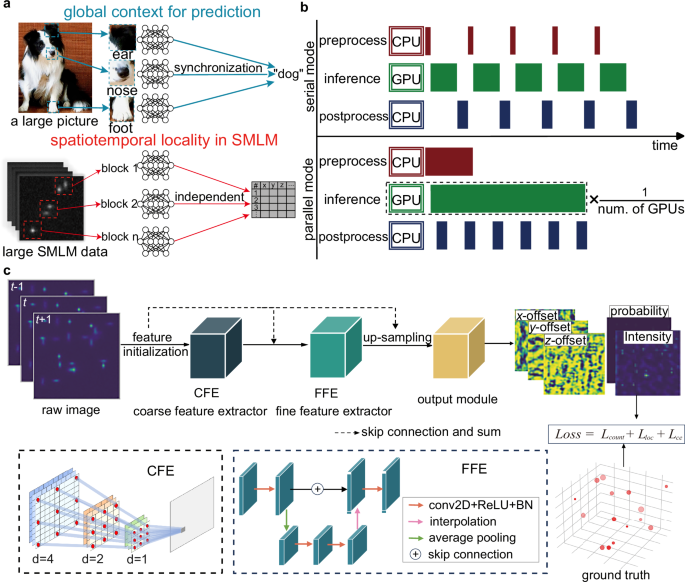
The scalable competitive SMLM (Single Molecule Localization Microscopy) data analysis framework, known as LiteLoc, offers impressive cross-platform compatibility, seamlessly functioning on Linux, Windows, and macOS operating systems. The entire data analysis pipeline comprises three key stages: preprocessing, inference, and postprocessing.
To harness the full potential of GPU resources and leverage multi-core CPU capabilities, LiteLoc employs a multi-process architecture. This design includes three parallel processes: a data loader for file reading and preprocessing, an analyzer for network inference, and a saver for result postprocessing and file writing. These components communicate efficiently using shared memory queues, allowing them to generate and retrieve data independently, thus maximizing throughput.
Data Preprocessing with the Data Loader Process
The data loader is the first process in the framework, tasked with reading SMLM files and conducting preprocessing tasks. Initially, it scans the designated data folder and formulates a reading plan that processes data in blocks—this strategy allows users to define block size, mitigating CPU memory constraints.
Once a block is loaded into the CPU, the data loader adds an extra frame at both ends and divides it into batches with a temporal overlap of two frames. This process ensures that each frame analyzed has a comprehensive three-frame temporal context, crucial for accurate inference by LiteLoc.
When dealing with large images that could overwhelm GPU memory, the data loader utilizes a divide-and-conquer method, crafting smaller overlapping mini-FOV (Field Of View) images akin to techniques employed in prior studies. This division enables the seamless feeding of spatiotemporally split batches, along with essential metadata, into the batch queue. By employing shared memory mechanisms via Python’s multiprocessing library, this strategy optimizes memory utilization and reduces communication overhead among processes.
Inference with Competitive Analyzer Processes
While the data loader prepares the input, the framework simultaneously activates multiple analyzer processes, each linked to a distinct GPU. This data-parallel strategy allows each GPU to possess its copy of the LiteLoc network, performing inference independently without needing synchronization.
This unique approach means that each analyzer can compete for data from the batch queue, overcoming potential bottlenecks caused by discrepancies in GPU processing speeds. Once inference is concluded, analyzers output multi-channel predictions, which undergo additional postprocessing steps. Notably, the framework also leverages GPU resources for the extraction of molecule lists from network predictions, making the transition from inference to detection smoother and more efficient.
Postprocessing with the Saver Process
The saver process plays a critical role in monitoring the result queue. It systematically retrieves intermediate results corresponding to batches of spatially overlapping data and performs essential postprocessing tasks. These tasks include aggregating results into their respective FOV positions, filtering out spatial overlaps, converting units, and preparing data for storage. The organized outputs are persistently written to a file defined by the user, ensuring that results remain easily accessible.
Architecture of LiteLoc
Central to LiteLoc’s functionality is its architecture, which consists of a Coarse Feature Extractor (CFE), a Fine Feature Extractor (FFE), and an Output Module. Each convolutional layer, except for those in the output module, is succeeded by a Rectified Linear Unit (ReLU) and batch normalization—fostering efficient training and feature extraction.
The initial input comprises three consecutive images analyzed independently by the CFE, generating down-sampled features through convolution and max pooling. Distinct from conventional methods, LiteLoc integrates dilated convolutions with exponentially increasing dilation factors, allowing for adaptive multi-scale feature extraction without losing spatial resolution.
Recognizing the necessity for capturing intricate patterns, LiteLoc employs a simplified U-Net structure, enhancing feature learning through strategic down-sampling and up-sampling. Neighborhood connections among layers facilitate the reuse of features, combatting gradient vanishing issues commonly encountered in deep networks.
The output module transforms processed features into final outputs, delivering pixel-wise probability maps indicating the presence of emitters, along with corresponding offsets and uncertainties. A dual-threshold strategy ensures robust detection even in challenging areas, enhancing localization accuracy.
GPU Cluster Configurations for Enhanced Throughput
To further amplify the data analysis capabilities, the framework is optimized for use with a powerful multi-GPU server housed in a compact 9U computer rack. Equipped with dual Intel Xeon Platinum 8375C CPUs and a staggering eight NVIDIA GeForce RTX 4090 GPUs, the server efficiently balances computational workload.
Running on Ubuntu 22.04 LTS and utilizing CUDA 12.4 alongside cuDNN, the server is fine-tuned for peak performance with PyTorch 2.3. A fast 4TB NVMe SSD ensures rapid data access, while advanced cooling systems maintain optimal operation temperatures, laying the foundation for efficient, large-scale data processing.
Optical Setups for Enhanced Imaging
The LiteLoc framework integrates a custom-built microscope, drawing from previously established designs. The optical configuration involves sophisticated setups for excitation light, including advanced fiber couplings and tunable illumination angles. By employing bandpass filters and deformations in optical paths, the microscope ensures precise light management, which is vital for achieving high-resolution imaging.
A closed-loop focus lock control system guarantees stability throughout the imaging process, employing a quadrant photodiode and z-piezo stage to maintain focal integrity in dynamic biological samples.
Biological Sample Preparation for Imaging
The implementation of LiteLoc begins with meticulous biological sample preparation, emphasizing cellular integrity and clarity. U2OS cells are cultivated under optimal conditions on specially cleaned glass coverslips, crucial for super-resolution microscopy.
For molecular labeling, the Nup96 protein is processed through a series of fixation and permeabilization steps, ensuring reliable bonding with fluorescent markers. This careful approach maximizes signal quality and consistency, which is vital for the accurate interpretation of super-resolution imaging results.
Innovative Imaging Buffer Composition
In imaging, utilizing a refractive index-matching buffer not only enhances visualization but also stabilizes biological samples. The buffer’s composition includes essential components like Tris-HCl, glucose oxidase, and thiodiethanol, guaranteeing a supportive environment for precise imaging under challenging conditions.
By integrating these intricate processes, protocols, and technologies, LiteLoc positions itself as a forefront solution for challenging data analysis in SMLM, enhancing both throughput and accuracy, essential qualities in cutting-edge biological research.