Investigating T-Cell Receptor Repertoires in Breast Cancer Patients
In a groundbreaking study, researchers delved into the intricate relationship between T-cell receptor (TCR) repertoires in peripheral blood and tumor status by comparing blood samples from women with breast cancer (BC) and healthy donors (HD). By integrating machine learning techniques, the study aimed to develop a classification model capable of accurately identifying the clinical status of previously unseen samples. This cutting-edge work not only sheds light on the potential of TCR profiles in cancer diagnostics but also paves the way for innovative approaches to immunotherapy.
The Research Methodology
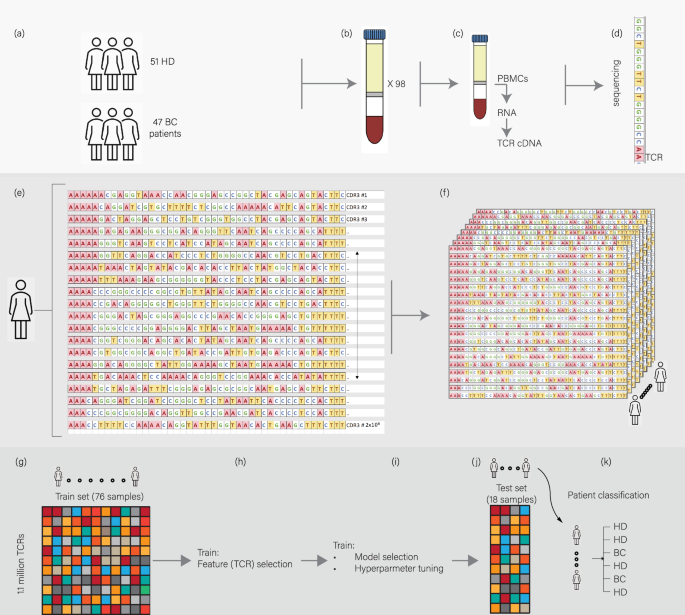
The study involved a structured methodology that encompassed several crucial steps, starting with the collection of samples. A total of 98 women participated; 47 diagnosed with breast cancer and 51 without any known health issues. Importantly, all samples from BC patients were collected before any treatment commenced.
Once collected, peripheral blood mononuclear cells (PBMCs) were isolated, from which RNA was extracted to create TCR-sequencing libraries. The libraries underwent high-throughput sequencing, resulting in a comprehensive characterization of TCR repertoires for each participant. The profound distribution of clones and their abundance was meticulously organized into a large data table, representing individual clones across patients and forming the basis for downstream analysis.
The pivotal step involved applying machine learning algorithms to this extensive dataset. Through feature selection and model training, researchers developed a robust classification framework aimed at differentiating between blood samples of BC patients and HDs.
Quality Control Assessments
To ensure data quality and comparability, several key repertoire characteristics were meticulously examined. Notably, the proportion of non-productive sequences (around 0.09) aligned with previously published datasets, confirming the reliability of the sequencing and annotation processes. Data visualization techniques, including heatmaps, illustrated consistent patterns of V and J gene usage, showing uniform distribution across both BC and HD groups.
The analysis also delved into CDR3 length distributions, revealing identical profiles in both cohorts, supporting the robustness of the dataset. Importantly, these quality control measures aimed to minimize potential technical artifacts, ensuring that the features employed in machine learning analyses remained reliable.
Analyzing Clonotype Quantification and Diversity
The initial analyses quantified unique clonotypes across all samples. Strikingly, the study found statistically significant differences between BC and HD groups, especially concerning the number of unique sequences and rare clonotypes—laying a foundation for distinguishing between the two populations. However, no considerable differences emerged among the most abundant clonotypes, highlighting nuances in TCR distribution.
Moreover, researchers assessed diversity metrics such as Gini diversity, the Inverse Simpson Index, and true diversity. Results demonstrated statistically significant variations between groups. Interestingly, despite these findings, the differences were insufficient for effective sample classification—a critical goal behind the research endeavor.
HLA Distribution Analysis
To rule out the human leukocyte antigen (HLA) profile as a confounding factor, researchers applied HLAGuessr to the dataset. The exploration revealed no significant differences in predicted HLA allele distributions between BC and HD groups, suggesting that HLA variations did not contribute to the observed TCR clonotype patterns.
Supervised Machine Learning Approach
The challenges inherent in traditional statistical methods prompted the adoption of supervised machine learning techniques. The dataset encompassed over one million rows and 94 columns, where each column represented a sample, and rows captured individual TCR sequence frequencies. The machine learning framework involved multiple rounds of subsampling and rigorous validation processes.
Notably, the training set was divided into subtraining and validation sets, with models evaluated based on predictive capabilities. A highly iterative process ensued, leading to the selection of three robust boosting models—AdaBoost, Gradient Boosting Machine, and XGBoost—as the methodologies demonstrated remarkable consistency across multiple iterations.
Ultimately, the XGBoost model was chosen for its superior performance, which achieved an area under the curve (AUC) score of 1.0 on the validation set and 0.96 on the test set.
V(D)J Combinations and T-Cell Subtype Identification
Identifying distinct T-cell subtypes hinges on analyzing unique V(D)J combinations characteristic of their specific TCR sequences. Following the feature selection protocol, the study pinpointed ten CDR3 sequences linked with specific T-cell types, including T cells associated with historically significant immune functions.
Database Analysis of Clonal Frequency
The analysis also harnessed external databases to investigate the frequency of identified clones. In databases such as VDJdb and McPAS, researchers found significant entries for certain identified TCRs, enabling a deeper understanding of their potential roles in cancer immunity.
TCR Embedding and Visualization
Lastly, the researchers utilized self-supervised learning for TCR embeddings, which facilitated visualization through UMAP plots. These plots revealed distinct clusters of TCR sequences, indicating that the classification model successfully captures meaningful patterns within the complex TCR repertoire.
This meticulous investigative approach not only enhances our understanding of TCR repertoires in breast cancer but also establishes a framework for future research into T-cell responses in various malignancies, potentially revolutionizing diagnostics and therapeutic strategies in oncology.