Ethical Considerations in Research: Compliance, Consent, and Integrity
Ethics Statement
In modern research, adherence to ethical standards is paramount. This ensures that all protocols align with established regulations that protect both participants and the integrity of the research process. The current study has been rigorously evaluated and approved by the Medical Science Research Ethics Committee of the First Affiliated Hospital of China Medical University, under the ethical code “kelunshen [2021] 2020-196-2” (number: AF-SOP-07-1.1-01). Notably, the research has been carefully designed to avoid including any personally identifiable information, affirming a commitment to confidentiality and privacy.
Informed Consent
Central to ethical research practices is obtaining informed consent from participants. In this study, consent was explicitly secured from the five pathologists involved in the slide-reviewing phase. Their willingness has been formally acknowledged by including them as co-authors on the manuscript, thereby recognizing their substantial contributions and ensuring transparency in authorship.
Data Acquisition and Preprocessing
Overview of the Dataset
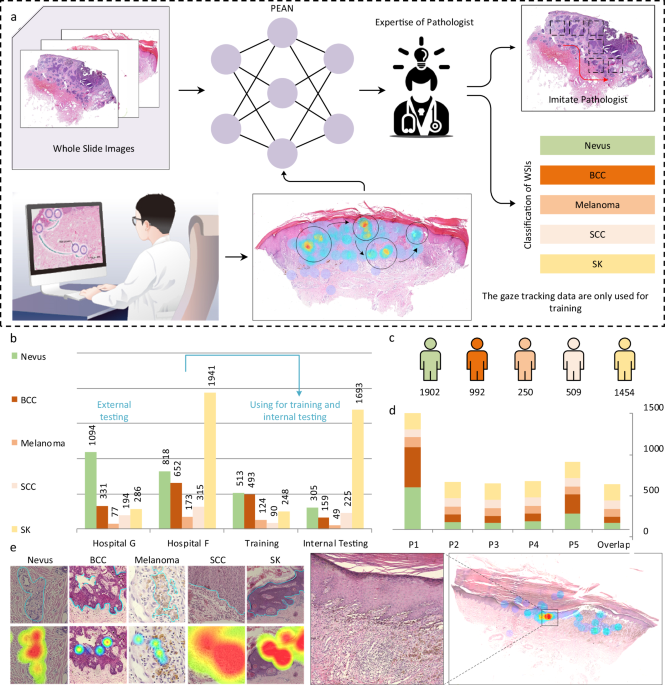
Before delving into the specifics of the deep learning model employed in this research, it is essential to understand the eye-tracking data set utilized. The dataset comprises 5,881 Hematoxylin and Eosin (H&E) stained whole slide images (WSIs) derived from four specific diseases—Basal Cell Carcinoma (BCC), Melanoma, Squamous Cell Carcinoma (SCC), and Seborrheic Keratosis (SK)—alongside images of a benign condition, nevus. The WSIs were meticulously collected from 5,107 patients over a six-year span (2016-2022) from the First Affiliated Hospital of China Medical University and the Shenyang Military Region General Hospital.
According to Supplementary Figure 2, the data collection was continuous to reflect a distribution representative of real-world pathology cases. However, some images were excluded—specifically, those of nevus and BCC that exhibited poor quality—following a re-examination process by the pathologists. Their evaluation criteria included assessing the adequacy of tissue samples, image clarity, and the presence of obstructions or artifacts. Only those WSIs subjected to unanimous approval from two pathologists were incorporated into the dataset.
Re-examination of Labels
Importantly, prior pathology reports associated with each WSI were also scrutinized to address potential inaccuracies. Instead of relying solely on these reports, the study employed a systematic labeling approach, particularly for cases with differing evaluations from the pathologists. In such instances, an additional senior pathologist was consulted to achieve a consensus on the final labeling.
Eye-Tracking Methodology
To collect eye-tracking data, the research team developed the “EasyPathology” software. This tool allowed pathologists to review the WSIs while simultaneously capturing their eye movement patterns using a Tobii Pro Spectrum eye tracker. The tracker records eye movements at a sampling frequency of 60Hz, mapping gaze locations onto the 2D WSI. Given the often-limited display size of individual WSIs, the software implements a “sliding window operation” to facilitate WSI review at varying magnifications.
The experimental setup was carefully designed to ensure data accuracy without disrupting the pathologists’ workflow. This included creating a soundproof environment, ensuring even lighting, and utilizing specified hardware configurations to optimize usability. Prior to data collection, the pathologists underwent training on how to use the EasyPathology software and eye tracker, with each review session starting with a calibration period.
Data Recording Strategies
Throughout the review process, the software recorded the pathologists’ activities, encompassing various aspects such as zooming, navigation, observation points, and final diagnoses while concealing the true labels to maintain impartiality. As pathologists reviewed WSIs for extensive periods, it was found that specific visual behavior features strongly correlated with diagnostic accuracy, leading to the establishment of 50 minutes as the ideal review duration.
Subsequent analysis of the eye-tracking data involved a color-coded approach to identify and exclude any instances of poor integrity, significant drift in eye movement, or operational errors. A thorough cleaning process ensured that the final dataset consisted of 3,978 slide-review sessions, with 542 WSIs reviewed by all participating pathologists.
Methodological Deep Dive: Eye-Tracking Data Processing
Preprocessing Steps
The eye-tracking data underwent meticulous preprocessing, which comprised the identification of observation points, the exclusion of non-meaningful visual behaviors, and the classification of pathologists’ visual behaviors. By employing Density-Based Spatial Clustering of Applications with Noise (DBSCAN), researchers differentiated between fixation and search behaviors based on observation point density.
Expertise Extraction
The extraction of pathologists’ expertise was framed through attention scores, captured while they manually reviewed the WSIs. The methodology aimed to model the probability distribution of the paths traced by expert pathologists. This entailed utilizing feature vectors extracted from images, with a focus on wounds observed at different magnifications.
An important notion underpinning the model was the principle of maximum entropy, which posits that the behaviors observed are nearly optimal responses to an underlying cost function. Various neural networks and image features were integrated to facilitate the analysis of the pathologists’ attention, leading to the extraction of a “pathology expertise value” for various image patches.
Reinforcement Learning Model
In a novel approach, a reinforcement learning (RL) model was constructed to emulate the visual behavior of expert pathologists during image reviews. Utilizing expert-generated trajectories, the RL framework analyzed sequential decision-making processes, empowering the model to effectively navigate WSIs and emulate the pathologists’ search patterns.
Conclusion and Future Directions
While this article doesn’t provide a concluding statement, it emphasizes ethical compliance, meticulous data acquisition, and innovative methodologies in this influential research. The emphasis on informed consent, careful data preprocessing, and systematic analysis showcases a robust commitment to both ethical standards and scientific integrity, paving the way for future studies in the realm of digital pathology and deep learning applications. By continuing to iterate on these methodologies, researchers can unlock further insights that enhance diagnostic accuracy and efficiency in pathology.