Unveiling the Power of Machine Learning in MRI Analysis: A Study from Penn Medicine BioBank
Introduction to the Research Context
In the evolving landscape of medical research, the utilization of machine learning technologies has revolutionized the analysis of complex imaging data. The recent study conducted using data from the University of Pennsylvania Health System exemplifies this trend, particularly through its association with the Penn Medicine BioBank (PMBB). The PMBB serves as a vital resource, providing a wealth of clinical information from participants who have consented to the use of their data for research endeavors. Under the oversight of an Institutional Review Board (IRB) and supported by notable contributions, including a generous endowment from the Smilow family and NIH funding, the PMBB operates within rigorous ethical guidelines.
An Overview of the Dataset
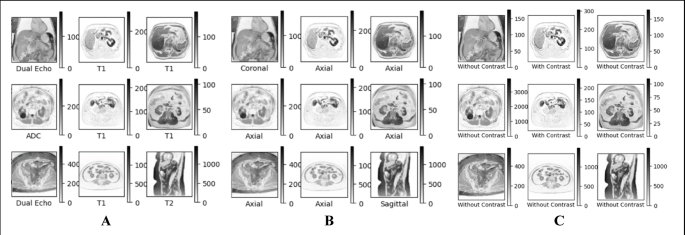
The dataset analyzed in this study comprises 26 unique examinations, correlating to various subjects. An impressive 576 series of abdominal MRI scans were collated, resulting in a staggering total of 29,484 2D images. However, quality control processes streamlined the dataset to 29,361 functional images, as corrupt DICOM files were discarded. Each MRI series is categorized based on several characteristics: 11 distinct pulse sequences such as ADC and DWI, orientations like axial or coronal, and binary contrast labels indicating whether intravenous contrast was used.
Data Characterization
This categorization is meticulously performed, with each MRI series verified manually by a board-certified abdominal radiologist to ensure accuracy. Each category contributes to a robust framework for analysis, ultimately enriching the dataset and enhancing the capacity to draw meaningful conclusions.
The Technical Framework
The analysis harnesses the power of modern programming tools and libraries, including Python 3.10 and platforms like Google Colaboratory. The researchers employed popular libraries such as Pydicom, Pandas, NumPy, and TensorFlow to execute a series of well-defined workflows—beginning with preprocessing of the DICOM-formatted images.
Data Preprocessing and Classifier Development
The preprocessing phase was pivotal in ensuring that only usable pixel data was retained, adhering closely to quality control protocols. Images underwent standardization to a uniform size of 128×128 pixels before being converted to grayscale—a critical step that facilitates effective classification later on.
The initial classifier development involved building histogram-based models to establish baseline performance metrics. These classifiers computed intensity histograms from the MRI slices, forming fixed-length feature vectors that characterized the global intensity profiles. A random forest classifier was then employed on these features to establish benchmarks against which deep learning models would later be evaluated.
Building Deep Learning Models
The cornerstone of the analysis involved training custom convolutional neural networks (CNNs), designed to accurately classify the MRI images based on their pulse sequence, orientation, and contrast type. The architecture of these models was crafted to capture intricate features residing within the MRI data, enabling them to discern between multiple classes effectively.
Training the Models
The training phase utilized the Adam optimizer and monitored accuracy using categorical cross-entropy as the loss function. A meticulous approach was adopted to avoid overfitting, which included implementing early stopping protocols predicated on validation loss assessments. The models were fine-tuned through five-fold stratified cross-validation within the training set, ensuring robustness in performance across various patient distributions. The results of this process yielded high-performing models that were subsequently evaluated against a wholly reserved test set.
Evaluation and Model Testing
As the models progressed to testing, they demonstrated remarkable proficiency in assessing the various attributes within the MRI images. Quantitative metrics such as accuracy, precision, recall, and F1 score were computed, revealing insights into the models’ predictive capabilities. A visual confusion matrix further illustrated the performance, depicting how frequently the models accurately identified classifications.
Understandability Through Grad-CAM
In an effort to demystify how models reach their conclusions, the researchers implemented Gradient-weighted Class Activation Mapping (Grad-CAM). This technique afforded vital transparency by generating heatmaps, which spotlighted the image areas most integral to the model’s predictions—an essential feature for the practical application of deep learning in clinical settings.
Majority Voting for Volume-Level Classification
To ensure model evaluations adequately reflected clinical practice, a majority voting mechanism was utilized for classifying MRI volumes. Each slice-level prediction was aggregated at the volume level, thereby aligning the model’s outputs with the interpretative practices of radiologists, who typically analyze imaging data holistically rather than in isolation.
External Validation with the Duke Liver Dataset
To test the generalizability of these machine learning models, the study engaged with an external dataset—the Duke Liver Dataset. This data comprises 2,146 abdominal MRI series from a diverse set of patients. The classification models were rigorously evaluated against this dataset, with effective mappings established from prior classifications.
Through strategic engagement with another institution’s dataset, the researchers sought to affirm the robustness and applicability of their models in varied clinical contexts. An illustrated comparison of series across different datasets emphasized the models’ adaptability in the face of new challenges.
The study exemplifies a profound integration of advanced technology in medicine, underscoring the dynamic potential for machine learning frameworks to augment diagnostic processes in radiology. As research continues in this space, insights from this study not only pave the way for enhanced imaging diagnostics but also bolster the infrastructures of medical data utilization.