AI-Driven Diagnosis of Malaria Using Blood Film Microscopic Images
The fight against malaria, a disease responsible for millions of infections globally, has seen a significant advancement with the advent of artificial intelligence (AI). This article details a robust AI-driven model designed to accurately diagnose malaria using microscopic images of blood films. The proposed methodology leverages a multi-phase approach to enhance the precision and reliability of the diagnostic process. This structured workflow incorporates several critical stages, including data preprocessing, feature extraction, model training, validation, and testing, all aiming to improve diagnostic accuracy and efficiency in both clinical and field settings.
Dataset Overview
The foundation of this solution is the “Cell Images for Detecting Malaria” dataset, available on Kaggle. This dataset contains a meticulously curated collection of 27,558 microscopic images, evenly split between two categories: parasitized (malaria-infected) and uninfected (healthy). Captured using light microscopy at a 100x magnification, these images maintain a high resolution crucial for machine learning applications. The original image size is 150×150 pixels, accommodating the proposed AI model’s input requirements following preprocessing steps, such as resizing.
The dataset is organized into two distinct directories: one for parasitized cells featuring irregular shapes and dark spots, and another for uninfected cells, marked by uniform shapes and smooth textures. This organization simplifies data loading and preprocessing tasks, enhancing the efficiency of model training. Importantly, the dataset is balanced in terms of class distribution, significantly minimizing potential biases that could arise during model development.
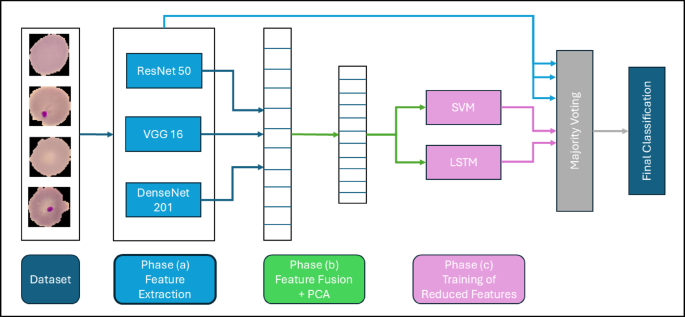
Phase (a): Implementation of End-to-End Deep Learning Models
The methodology employs modern deep learning techniques by utilizing three state-of-the-art convolutional neural network (CNN) architectures: ResNet-50, VGG-16, and DenseNet-201. These models act as feature extractors, drawing on pre-trained weights derived from large-scale datasets to recognize intricate patterns in blood cell images.
-
ResNet-50: This model can capture 2048 deep hierarchical features through its unique residual connections, which effectively tackle vanishing gradient problems.
-
VGG-16: Known for its straightforward structure, VGG-16 identifies 4096 spatial features using uniform convolutional layers, which ensures consistent detail extraction.
- DenseNet-201: DenseNet enhances feature propagation by establishing densely connected layers, thereby enabling efficient feature reuse.
These architectures are not only responsible for feature extraction but are also further fine-tuned as end-to-end models specifically tailored for malaria diagnosis. This dual approach enables the learning of both low-level features and high-level concepts relevant to identifying infected cells, establishing a strong foundation for accurate diagnosis.
Phase (b): Feature Fusion and Dimensionality Reduction
After the independent feature extraction phases, we enter feature fusion, synthesizing outputs from ResNet-50, VGG-16, and DenseNet-201 into a consolidated feature vector. The concatenation takes the form:
[
F{fused} = [F{ResNet}, F{VGG16}, F{DenseNet}]
]
However, this fusion introduces high dimensionality, which can complicate computations and increase the risk of overfitting. To address this, Principal Component Analysis (PCA) is applied. PCA identifies the principal components that maximize variability in the feature set, condensing the dimension of the dataset while preserving critical information.
[
F{PCA} = F{fused} \cdot W
]
PCA aids in retaining only the most significant features while mitigating computational burdens. This step serves to accelerate subsequent classification tasks while maintaining diagnostic accuracy.
Phase (c): Hybrid Classification Framework
Post-dimensionality reduction, the refined feature vectors proceed into a hybrid classification framework combining traditional machine learning with deep learning approaches. This framework comprises two core components: Support Vector Machine (SVM) and Long Short-Term Memory networks (LSTM), both integral to achieving robust and accurate classification.
Support Vector Machine (SVM)
SVM is recognized for its ability to delineate optimal decision boundaries between classes, even within high-dimensional contexts. It identifies hyperplanes that best separate the two classes (malaria-infected vs. healthy). The SVM approach maximizes the margin, enhancing generalization to unseen data and maintaining classification robustness.
The mathematical representation for SVM finds the optimal hyperplane as follows:
[
f(x) = w^T x + b = 0
]
where ( w ) signifies the weight vector and ( b ) represents the bias. It aims to minimize the following optimization problem:
[
\underset{w, b}{min} \frac{1}{2}|w|^2
]
with constraints ensuring correct classification of each training instance.
Long Short-Term Memory Networks (LSTM)
LSTMs extend natural language processing capabilities into the realm of image classification. While the feature vectors are not sequential, LSTMs excel at capturing contextual relationships among features. LSTMs possess unique gates controlling information flow, enabling the model to retain essential patterns while discarding irrelevant noise.
The intricate architecture of LSTM cells comprises a forget gate, input gate, and output gate, facilitating the retention of long-term dependencies crucial for pattern recognition in biological data. Their application in malaria diagnosis enriches the interpretative capacity of the classification pipeline.
Decision Aggregation
To enhance the reliability of the classification process, a majority voting mechanism aggregates predictions from all classifiers involved, including the three deep learning models (ResNet-50, VGG-16, DenseNet-201), alongside SVM and LSTM. Each model casts a vote on whether the blood film image indicates malaria infection or not.
This democratic approach capitalizes on the strengths of diverse models, reducing biases from any individual classifier. Through this consensus-building strategy, the diagnostic system can confidently and accurately determine the likelihood of malaria infection—an essential attribute for deployment in real-world clinical settings.
The carefully constructed workflow encompassing dataset preparation, feature extraction, dimensionality reduction, hybrid classification, and decision aggregation encapsulates a holistic approach to malaria diagnosis, elevating AI’s potential in the medical domain.