Understanding the Methods and Processes in Pain Detection Research
This article delves into the critical methods and processes utilized in the study of automated pain recognition, focusing primarily on two significant datasets: the BioVid Heat Pain Database and the AI4PAIN Database. This exploration encompasses the evaluation methodologies, data preprocessing techniques, and deep learning architectures employed to create an advanced framework for pain detection.
BioVid Heat Pain Database
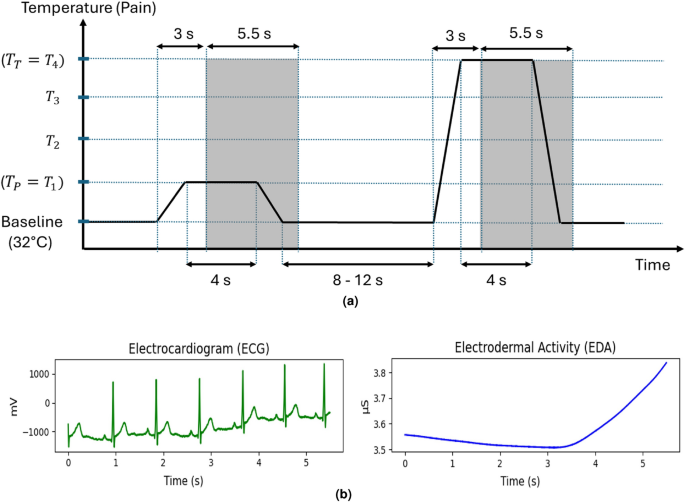
The BioVid Heat Pain Database is a well-established resource in the field of pain recognition studies. For this research, we utilized two specific physiological modalities: Electrodermal Activity (EDA) and Electrocardiography (ECG) data. The dataset boasts recordings from 87 healthy participants, comprising 44 males and 43 females across three age groups: 20–35 years, 36–50 years, and 51–65 years.
Each participant underwent controlled heat pain stimuli on the right arm, facilitated by a thermode. Four subject-specific pain intensities were tested, with 20 repetitions across all levels of pain. Each stimulus lasted four seconds, followed by a random resting state of 8 to 12 seconds, capturing a variety of physiological responses.
Despite its extensive use in various studies, the BioVid database poses some challenges regarding the comparability of results. One significant limitation arises from the inconsistency in the selection of subjects. Some studies have chosen to exclude participants who displayed minimal reactions to the pain stimulus, thus potentially skewing the dataset. Furthermore, variations in analysis timing add another layer of complexity, as peak responses to pain often occur several seconds after stimuli initiation.
In this study, we retained all available data for a more comprehensive analysis of pain responses while ensuring our comparisons were made with studies employing similar datasets.
AI4PAIN Dataset
The AI4PAIN 2025 Grand Challenge Dataset represents the forefront of resources designed to enhance research capabilities in automatic pain recognition. It includes physiological recordings divided into four biosignal modalities: EDA, Blood Volume Pulse (BVP), Respiration (RESP), and Peripheral Oxygen Saturation (SpO2). The data acquisition employed Biosignal Plux sensors, maintaining a sampling rate of 100 Hz, and extended across 65 participants aged 17 to 52.
Pain stimuli were administered using a transcutaneous electrical nerve stimulation (TENS) device, which aimed to induce low to high pain responses. A structured protocol involving a baseline measurement and multiple pain-inducing trials was implemented. Notably, for our framework evaluation, we selected only the EDA and BVP signals, using BVP instead of ECG to maintain relevance while adhering to the dataset’s features.
Data Preprocessing Techniques
A robust preprocessing pipeline is crucial for managing the complexities of physiological data. The BioVid database includes both raw and preprocessed signals; for our experiments, we opted for the preprocessed version. EDA signals underwent filtering through a Butterworth low-pass filter, while ECG signals were processed using a Butterworth band-pass filter designed for the 0.1 to 250 Hz frequency range. To ensure uniformity, all physiological signals were downsampled to 25 Hz, allowing for consistent input dimensions across trials.
On the flip side, the AI4PAIN dataset was utilized in its raw form. This provided a unique opportunity to assess the proposed method’s performance on unfiltered data, thus evaluating its robustness in handling various input conditions.
Proposed Framework for Pain Detection
The heart of our research lies in the Cross-Mod Transformer, a deep learning framework specifically designed for pain recognition through physiological data. This model incorporates a hierarchical fusion mechanism, combining fully convolutional networks (FCN), attention-based long short-term memory (ALSTM) networks, and Transformers.
The architecture is organized into two main streams: Uni-modal Stream and Multi-modal Stream.
Uni-modal Stream
In the Uni-modal Stream, separate networks process each physiological signal independently to extract unique temporal patterns. Here, the FCN captures short-term features while the ALSTM focuses on long-term dependencies. The integration of these two models capitalizes on their strengths, with the FCN maintaining the temporal resolution and the ALSTM mitigating issues like vanishing gradients.
Multi-modal Stream
The Multi-modal Stream leverages a separate Transformer to fuse the two modalities—EDA and ECG. This stage emphasizes the inter-modal relationships between the signals, harnessing their complementary features for enhanced pain recognition. By focusing on the interactions between modalities, the framework aims to improve prediction accuracy significantly.
Experimental Setup
For our experiments, we utilized a high-performance system equipped with an NVIDIA GeForce RTX 4080 GPU and a 13th Gen Intel Core i7 CPU. The training procedure involved a multi-step approach, where individual networks were trained separately to avoid modality competition—an issue that can hinder performance if modalities interfere during joint training.
We adopted a rigorous evaluation process, utilizing 10-fold cross-validation and leave-one-subject-out methodologies with the BioVid dataset to ensure fair comparisons across studies. The evaluation metrics comprised accuracy, F1 score, sensitivity, and specificity, providing a comprehensive measure of model performance.
Additionally, the interpretability of the model was explored through visualizations of attention maps, allowing us to understand better how the model prioritizes various physiological signals during classification tasks.
This structured exploration of data and methodologies highlights the intricate processes essential for advancing pain recognition technologies, providing a deep insight into the challenges and innovations in this vital field of study.