Breakthrough: Deep Learning Enhances CRISPR Base-Editing Predictions with Multi-Dataset Training
Breakthrough: Deep Learning Enhances CRISPR Base-Editing Predictions with Multi-Dataset Training
Understanding CRISPR Base Editing
CRISPR base editing is a technique that allows scientists to modify specific DNA sequences without cutting both strands of the double helix. This refinement of classical CRISPR-Cas9 technology employs a modified Cas9 enzyme combined with a deaminase that chemically converts one DNA base into another. For example, adenine base editors convert A·T base pairs to G·C, while cytosine base editors convert C·G to T·A. The impact of this technology lies in its ability to make precise genetic edits, which is crucial for developing therapies for genetic disorders.
The Importance of Predictive Models
Predicting the outcomes and efficiency of CRISPR base editing remains a significant challenge. Existing predictive tools are often trained on various individual datasets, which may lead to limitations in their application. Each dataset can vary due to differences in experimental conditions, deaminase variants, and other influencing factors. For instance, some models may perform well in identical experimental setups but struggle to generalize to other conditions, which can slow down research progress in genetic engineering.
Key Components of the Study
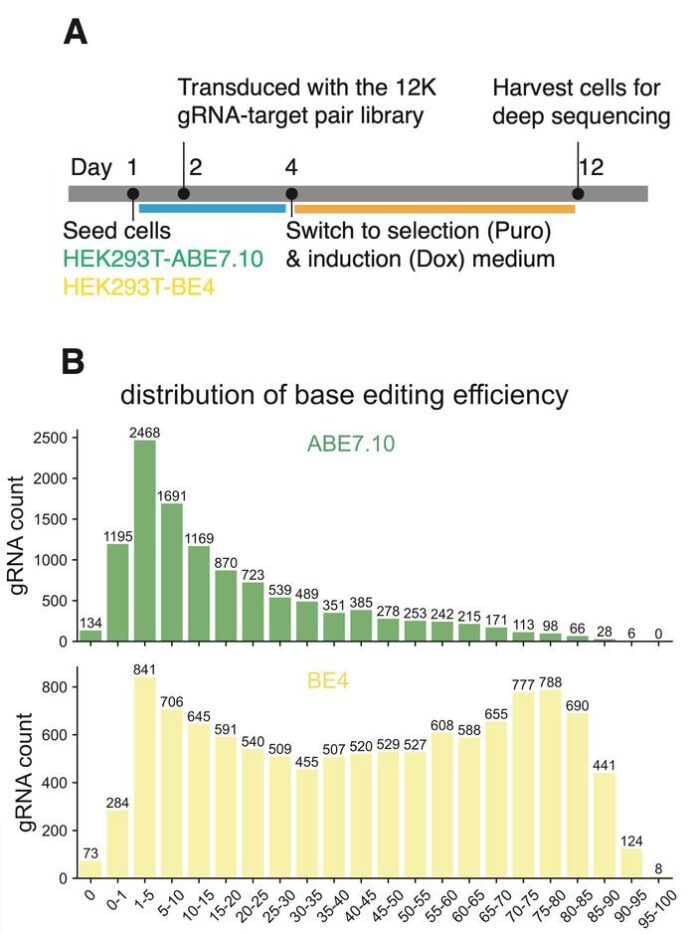
The researchers developed a new approach using dataset-labeled multi-dataset training to improve prediction models for CRISPR base-editing efficiency. They created a more robust dataset by generating new experimental data for approximately 11,500 gRNAs through their SURRO-seq technology. This technique allowed for a comprehensive analysis of base-editing efficiency for adenine and cytosine base editors in HEK293T cells. The research highlights how integrating different datasets—rather than treating them in isolation—enhances model training and accuracy.
The Step-by-Step Training Process
To develop the new predictive model, the authors employed a multi-stage process. First, they labeled each guide RNA (gRNA) based on its dataset of origin. Then, they created a deep learning architecture capable of simultaneously training on multiple datasets. They leveraged convolutional neural networks to evaluate target sequence characteristics and molecular features like gRNA-DNA binding energy. This stepwise integration of varying data is crucial as it allows the model to learn from a diverse range of experiments, improving its predictive capabilities.
A Practical Case Study
One striking outcome from the study was the superior predictive performance of the CRISPRon-ABE and CRISPRon-CBE models compared to existing methods. The metrics indicated that including dataset labels significantly improved the models’ accuracy—by about 10%—compared to models that did not account for these labels. This demonstrates a practical application where researchers can depend on these models to select the most efficient gRNAs for specific editing tasks, thereby optimizing their outcomes.
Common Mistakes and Solutions
A frequent mistake in employing CRISPR base editing techniques involves not taking into account the wide variability in dataset quality. Improper analysis can lead to inaccurate predictions, diminishing the effectiveness of genetic modifications. The solution lies in the new methodology, which explicitly incorporates dataset origin during model training, allowing researchers to assign weights to different datasets based on their unique conditions. This level of sophistication ensures higher fidelity in predictions.
Tools and Frameworks Available for Researchers
The study introduces CRISPRon-ABE and CRISPRon-CBE, which are now accessible as both a web server and standalone software. The tools allow researchers to input various parameters, including the target DNA sequence and dataset labels. Users can generate predictions that reflect the best possible gRNAs for their specific experimental contexts. This ecosystem of tools provides invaluable support for researchers engaged in genetic editing.
Alternatives and Their Pros and Cons
While CRISPRon-ABE and CRISPRon-CBE present significant advantages, alternative methods like DeepABE/CBE and BE-HIVE have their pros and cons. For instance, older models may require less computational power but often compromise on predictive accuracy when faced with diverse experimental settings. In contrast, the new models demand more resources but offer a higher degree of precision and reliability in predictions by accommodating multiple datasets.
FAQ
What data sources were used for training the model?
The research team used multiple datasets, including their newly generated SURRO-seq data and previously published datasets to enhance model accuracy.
How does the model handle data incompatibility?
The model labels each input’s dataset origin, allowing it to learn the differences in experimental conditions and effectively merge data during training.
Are there limitations to the current models?
Yes, the current models primarily include three base editor variants and are based on training data from HEK293T cells, which may affect predictions for other cell types.
How can I access the models?
The CRISPRon-ABE and CRISPRon-CBE models are available via a web server, as well as standalone software for academic use.