Data Acquisition Procedure in Rodent Sleep Research
Data acquisition is a fundamental process in scientific research, particularly in studies focused on understanding sleep dynamics in animal models. This article delves into the intricacies of the data acquisition procedure utilized in a recent study involving adult rats, shedding light on the methodologies, ethical considerations, and technical implementations necessary for robust sleep stage classification.
Cohorts and Ethical Considerations
In our study, we utilized three distinct cohorts encompassing a total of 36 unique adult male and female rats. The rats were divided into:
- Project Aurora dataset: 16 Wistar rats used for model training and validation.
- SleepyRat dataset: 14 Wistar rats for additional validation.
- SnoozyRat dataset: 6 Fischer 344/Brown Norway F1 hybrid rats for extensive examination.
All experiments adhered to the guidelines established by the Institutional Animal Care and Use Committee (IACUC) at the University of South Carolina, ensuring compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Further, the facility was fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). The rats were maintained on a 12-hour light-dark cycle to simulate natural conditions, critical for ensuring the validity of the sleep data acquired.
Implantation of EEG/EMG Transmitters
To gather accurate physiological data, rats underwent surgery to implant EEG/EMG transmitters (PhysioTel HD-S02). Using isoflurane anesthesia, the rats were affixed to a stereotaxic frame, where precise incisions were made into the abdominal region for intraperitoneal transmitter placement, and into the midline of the skull for EEG lead attachment.
Two EEG leads were securely positioned into stainless-steel screws, inserted into burr holes located at specific coordinates relative to bregma. Meanwhile, EMG leads were implanted in the cervical neck muscles, precisely distanced to ensure accurate measurement of muscle activity. After the surgical procedures, the skin was sutured, allowing for a recovery period of at least seven days before the initiation of sleep data acquisition sessions.
Sleep Data Acquisition Protocol
The primary sleep data collection took place in a designated quiet room where rats were undisturbed for two continuous 24-hour recording sessions. During these sessions, data was gathered using a PC running Ponemah 6.10 software (DSI), capturing EEG/EMG data at a sampling rate of 500 Hz. The digitized data were then exported in the European Data Format (EDF) for subsequent processing and analysis.
An expert in sleep staging manually annotated each recording in 10-second epochs utilizing NeuroScore 3.0 software. This expert scoring was pivotal in establishing the ground truth for our machine learning algorithms, facilitating robust training on the Project Aurora dataset. Detailed methodologies on training experts for manual sleep staging are available in our supplementary materials.
Data Structuring for Model Training
From the two recording sessions, we derived 16 discrete 24-hour EEG/EMG signals, one for each rat. We then combined these recordings into 48-hour sleep data sequences while preserving the identity of each rat throughout the dataset—ensuring that the training and evaluation of the model remained accurate.
The structure of our dataset, described as 16 rats x 48-hour PSG recordings, enabled effective modeling. This configuration was critical in training our neural network to generalize accurately across different input epochs while reducing potential biases that might emerge from data processing.
Validation Strategy
An integral aspect of model development is a rigorous validation strategy that simulates real-world deployment. We implemented a leave-one-out cross-validation method, where data from one rat formed a unique test fold while the remaining rats were used for training. This strategy is particularly beneficial, especially with limited data, as it promotes reliable generalization of our model beyond the training dataset.
Algorithm Design and Architecture
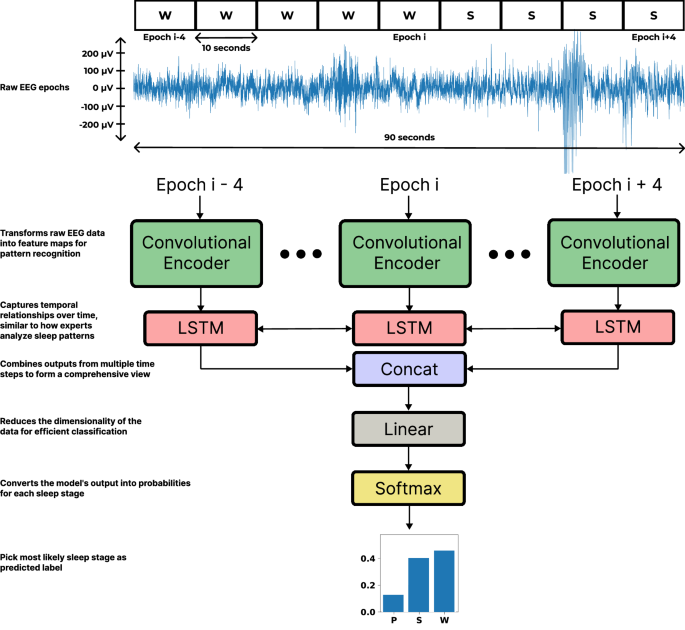
The model developed to classify sleep stages was designed strategically to encode EEG epochs into informative low-dimensional vectors before incorporating sequential information from neighboring epochs. A combined approach utilizing Convolutional Neural Networks (CNNs) alongside Long Short-Term Memory (LSTM) neural networks allowed us to achieve this objective effectively.
Each input sequence consisted of 9 ten-second epochs, predicting the sleep stage of the center epoch. The architecture of our model is detailed in accompanying figures and showcased its efficacy in tackling the inherent challenges of sleep stage classification from single-channel EEG data.
Epoch Encoder and Classifier
Our epoch encoder employed a modern variant of CNN known as RegNet, which facilitated optimal feature extraction through a series of residual blocks and global average pooling. The resulting feature sequences were processed by an LSTM, which integrated spatial and temporal information before encoding it into output states indicative of three vigilant states: REM sleep (P), NREM sleep (S), and wakefulness (W).
Preparation of Dataset for Training
For effective training of our model, input sequences were padded to allow for evaluation of the initial epochs while ensuring that the model could process continuous data streams without interruptions. The training protocol followed consisted of segments of 10-second epochs using a simple moving window approach, enabling us to construct a robust model that effectively distinguished between different sleep states.
Optimization Strategies Employed
Model training involved mini-batches of size 32, and the validation loss was monitored diligently to prevent overfitting through early stopping measures. Hyperparameter optimization was performed through a systematic grid search, leading us to establish an optimal configuration. The use of cross-entropy loss, combined with the Adam optimizer for its adaptive learning rates, proved advantageous in achieving stable convergence throughout training.
All processes were implemented within the Python environment utilizing PyTorch, taking advantage of GPU resources with NVIDIA RTX 4090 graphics cards to enhance performance and computational efficiency.
Defining Evaluation Metrics
Establishing meaningful evaluation metrics was crucial for validating our model’s performance, particularly in relation to the imbalanced classification nature of sleep staging. We primarily focused on the F1-score, a critical metric that balances precision and recall across multiple classes, thereby providing a nuanced view of our model’s efficacy in real-world applications.
Comparative Analysis with Previous Work
To validate our findings further, we compared our model’s performance against a recent sleep-staging algorithm developed by Liu et al. This comparative analysis utilized a comprehensive dataset (SPINDLE) encompassing diverse species, underscoring the generalizability and adaptability of our approach across various experimental contexts.
Validation of Conclusiveness on Vigilance State Duration
Finally, the SleepyRat dataset was employed to assess the conclusions drawn regarding vigilance state duration and architecture. An expert annotated the sleep stages for analysis, allowing us to perform statistical analyses and comparisons between human-annotated scoring and the model’s predictions.
In essence, the data acquisition procedures we’ve analyzed provide a foundational understanding of how rigorous methodology in animal studies can yield valuable insights into sleep dynamics and contribute to advancements in machine learning applications. Each step, from ethical considerations to state-of-the-art modeling strategies, plays a pivotal role in shaping the outcomes of such research, paving the way for future explorations in the realm of sleep science.