Study Design and Participants in Idiopathic Inflammatory Myopathy-Related Interstitial Lung Disease Research
Study Overview
This single-center, retrospective study was rigorously designed in accordance with the Helsinki Declaration. It received ethical clearance from the Human Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University, marked by approval number [KYLLSL201312202]. Notably, the Institutional Review Board waived the need for written informed consent since the study utilized anonymous clinical and imaging data.
Recruitment of Participants
Between January 2015 and August 2022, a total of 1,132 consecutive patients were examined. These individuals met the 2017 EULAR/ACR diagnostic criteria for idiopathic inflammatory myopathies (IIM) and were also evaluated against the diagnostic standards established during the 239th ENMC International Workshop. This included a comprehensive review by rheumatologists at our institution to reconfirm diagnoses.
Inclusion and Exclusion Criteria
For participants to be included in the study, they had to meet specific criteria:
- Confirmed diagnosis of Interstitial Lung Disease (ILD): A consensus imaging pattern diagnosis was achieved through a multidisciplinary discussion involving experts in ILD, radiology, and pathology.
- Availability of Chest High-Resolution Computed Tomography (HRCT): Quality imaging data was essential.
Exclusion criteria were equally rigorous:
- Inadequate HRCT Quality: Images lacking diagnostic clarity were omitted.
- Pulmonary Infection: Diagnoses confirmed through a multi-tiered approach including medical records, radiological assessments, and microbiological workups.
- Ambiguous HRCT Findings: Images that could not be classified into a specific pattern were excluded.
- Other Lung Diseases or Surgical History: Any signs of lung diseases unrelated to IIM-ILD or prior chest surgery disqualified participants.
Ultimately, 629 patients were included in the study cohort, with baseline HRCT images acquired during the initial diagnosis of ILD. These images were anonymized and stored in DICOM format for analysis.
Data Segmentation and Utilization
Data collected from January 2015 to December 2019, comprising 517 patients, was used for model development and internal testing. The following set of data, ranging from January 2020 to August 2022 with 112 patients, provided a basis for temporal external validation.
Dataset Organization
A stratified random sampling method was applied, dividing the former data into training and testing sets at a ratio of 7:3:
- Training Set: 361 subjects
- Internal Testing Set: 156 subjects
Each HRCT image’s patterns were reconfirmed by two thoracic radiologists, with assistance from a senior radiologist for challenging cases. The distribution of ILD imaging patterns within the training, testing, and validation datasets was as follows:
- Training Set: NSIP (136), OP (101), NSIP + OP (71), UIP (32), DAD (21)
- Testing Set: NSIP (59), OP (44), NSIP + OP (30), UIP (14), DAD (9)
- Validation Set: NSIP (53), OP (28), NSIP + OP (22), UIP (1), DAD (8)
Algorithms and Frameworks for Classification
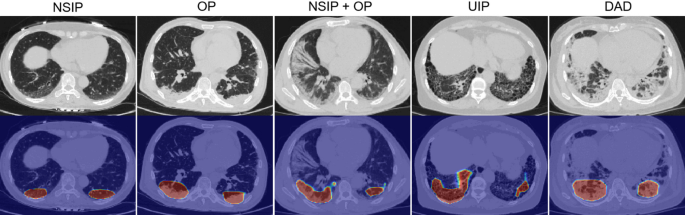
The study introduced a deep learning model designed to classify five categories of ILD imaging patterns, encompassing three critical components: image preprocessing, lung segmentation, and model evaluation.
Image Preprocessing
The imaging data underwent meticulous preprocessing:
- Conversion to Hounsfield Units (HU): This was essential for standardizing images.
- Resampling: All CT scans were standardized to a 1 x 1 x 1 mm slice thickness.
- Windowing and Normalization: By applying lung window adjustments and Z-score standardization, the CT images underwent a final normalization process targeting a uniform resolution and density range.
Lung Segmentation
Automated lung segmentation utilized the uAI Research Portal (Shanghai United Imaging Intelligence, Co., Ltd.), which deployed deep learning algorithms to focus specifically on lung abnormalities. Initial training reached impressive Dice similarity coefficients of 98.8% for lung parenchyma and 91.6% for abnormalities, with adjustments made as deemed necessary by thoracic radiologists.
Model Construction
To bolster the training set’s diversity and mitigate overfitting, a data augmentation process was applied, enabling random flips, scaling, and rotations of images. The core architecture was built on a Residual Network (ResNet), designed specifically for image recognition tasks.
The model incorporated class activation mapping (CAM), which aids interpretability by highlighting key areas of interest in the imaging data. Both label smoothing cross-entropy and mean squared error functions were utilized to guide network attention effectively.
Statistical Analysis Techniques
Data analyses employed scikit-learn and SciPy libraries, employing various statistical measures including:
- ANOVA and Chi-Square Tests: For baseline clinical comparisons between CT pattern subgroups.
- Classification Metrics: Model performance was evaluated across several dimensions, including accuracy, precision, specificity, sensitivity, and F1-score.
- Hamming Loss: This served as a metric for the model’s multi-class classification performance.
The results conveyed through these analyses were represented in mean ± standard deviation or as frequencies where applicable. Statistical significance was predetermined at a threshold of P < 0.05.
This comprehensive approach highlights not only the intricacies of the study design and participant selection but also the methodologies employed for data analysis, ensuring a robust examination of idiopathic inflammatory myopathies-related interstitial lung disease.