Evaluating Grad-CAM and Its Impact on Discriminator Performance for Histopathological Image Enhancement
When it comes to enhancing the quality of histopathological images, particularly those derived from frozen sections, the use of advanced methods such as Generative Adversarial Networks (GANs) has become increasingly prevalent. In our exploration, we focus on the Self-Attention Network (SAN) and how Grad-CAM serves as a powerful tool for evaluating its effectiveness, specifically in improving the quality of nuclei regions in these images.
Understanding Grad-CAM
Gradient-weighted Class Activation Mapping (Grad-CAM) is a visualization technique that sheds light on the decision-making process of neural networks. It does so by highlighting regions within an image that contribute most significantly to the model’s output, offering insights into how certain features influence the neural network’s decisions. This method proves invaluable in examining the performance of discriminators used in GAN architectures.
Enhancing Discriminator Focus: The Power of SAN
Our technique implements SAN to optimize the discriminator’s attention on the nuclei regions, which are crucial for accurate histopathological analysis. By analyzing the Grad-CAM outputs from the discriminator (D_P), we observed significant differences in how the various models (CycleGAN, CycleGAN with Attention U-Net & ResNet, and SAN) focus on the nuclei.
Visual Insights into Discriminator Attention
Figure 4 illustrates the Grad-CAM maps generated for different models. It becomes evident that the SAN method markedly enhances attention to the nuclei regions compared to other architectures. This improvement suggests that the generator is prompted to focus more on generating accurate nuclei content, which is essential for producing reliable histopathological images.
Frechet Inception Distance (FID) Score Evaluation
To quantitatively assess the similarity between the generated permanent section images and their unpaired real counterparts, we employed the Frechet Inception Distance (FID). This metric evaluates the Wasserstein distance between multivariate Gaussian distributions that represent the feature representations of both the generated and real images. A lower FID score indicates greater similarity, making this measure particularly useful for gauging enhancements in image quality.
Comparative Performance
In Table 3, FID scores for various methods—CycleGAN, CycleGAN with Attention U-Net & ResNet, and our SAN—reveal a consistent trend: the SAN method achieves lower FID scores across the test data from diverse tissue samples (breast, colon, and kidney cancers). This result underlines the superior image generation capabilities of our approach.
Visual Inspection of Generated Images
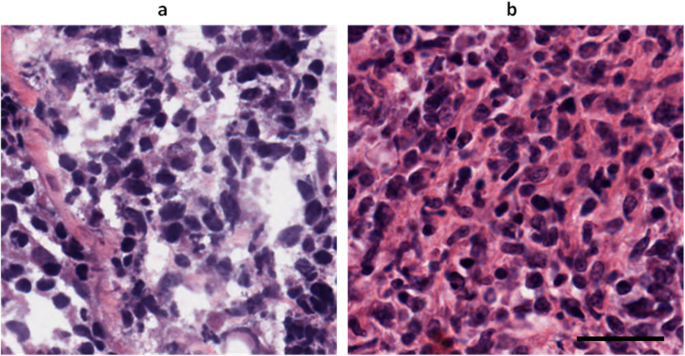
To further validate our model’s efficacy, we conducted a visual inspection of generated permanent section images. Figure 5 exemplifies how our SAN model successfully creates permanent section patches from frozen section patches while maintaining nuclei segmentation accuracy.
Quality Comparison
Figures comparing outputs from CycleGAN, CycleGAN with Attention U-Net & ResNet, and SAN—particularly for breast, colon, and kidney tissues—reveal some striking differences. While CycleGAN often produced images with noticeable artifacts and lacking details—especially in nuclei regions—our SAN approach demonstrated remarkable enhancement, yielding clearer, more diagnostically relevant images. The visual fidelity in contrast to frozen sections is crucial, as it directly supports improved diagnostic outcomes.
Nuclei Texture Analysis
A quantitative analysis of nuclei region textures was conducted using the Gray-Level Co-occurrence Matrix (GLCM). This analysis is pivotal for extracting essential texture features relevant to histopathological diagnostics. Utilizing 14 × 14-pixel crops focused solely on nuclei regions enables us to capture the most critical diagnostic details.
GLCM Feature Statistics
Table 4 summarizes the GLCM features across different samples, including permanent, frozen, CycleGAN-generated images, and SAN-enhanced images. Notably, the SAN method produced GLCM values closely aligning with those of real permanent sections, particularly in critical features like contrast and energy—key indicators of texture nuances essential for accurate diagnosis.
Jensen-Shannon Divergence Analysis
To establish texture similarity in a quantitative manner, we computed Jensen-Shannon (JS) divergence. Lower JS divergence values for SAN indicate closer alignment of generated textures with real permanent sections. As shown in Table 5, SAN method outperformed the other approaches in several key texture features, emphasizing its capacity to capture fine details critical for diagnostics.
Clinical Evaluation of SAN Enhancements
To assess the clinical utility of our SAN method, two independent pathologists evaluated matched histopathological image pairs from kidney tissues using established criteria. This validation aimed to determine how well SAN images enhanced diagnostic capabilities.
Pathological Assessment Findings
The evaluation revealed significant improvements across a range of critical morphological parameters, as summarized in Table 6. Parameters such as stain quality and nuclear morphological features scored exceptionally high, demonstrating SAN’s capability to enhance eosinophilic cytoplasm staining and reduce bubble artifacts. Its success in addressing these limitations directly correlates with improved diagnostic accuracy.
Detailed Diagnostic Significance
Improvements in nuclear detail and cellular architecture were evident, with high reliability observed in assessments of nuclear pleomorphism and cell membrane characteristics. High inter-pathologist agreement underscored the method’s potential to enhance intraoperative diagnostic accuracy due to the improved visualization of nuclear details.
Visual Comparison and Impact Analysis
A comprehensive assessment of 826 patches demonstrated substantial diagnostic improvements across numerous parameters, with significant enhancements in nuclear detail, cytoplasmic clarity, and cell border definition. This quantitative analysis underscores SAN’s potential to address critical diagnostic limitations inherent in frozen sections, enhancing the reliability of rapid intraoperative diagnoses.
Overlay Visuals
Finally, Figure 9 visually encapsulates the enhancements achieved by the SAN method, with overlays comparing frozen and SAN-enhanced images. This side-by-side comparison accentuates the significant advances made in nuclear detail, clarity, and tissue architecture preservation, crucial for ensuring high-quality diagnostic outcomes.
Through the integration of powerful tools like Grad-CAM alongside sophisticated models like SAN, we find ourselves at the forefront of histopathological image enhancement, paving the way for more accurate and reliable diagnostic practices in clinical settings.