Exploring the FANTASIA Pipeline: A Step-by-Step Guide
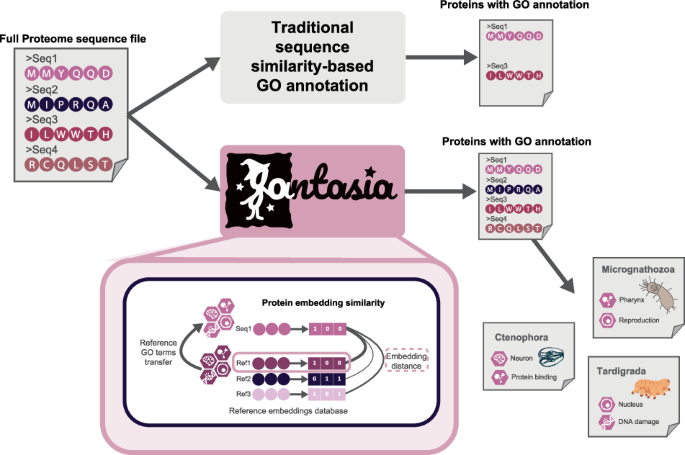
FANTASIA (Functionally ANnoTating All Proteomes Using Protein Embedding Similarity Analysis) is a cutting-edge reimplementation of the original GOPredSim algorithm designed for functional annotation of full proteomes. This enhanced iteration improves usability, scalability, and installation reliability, catering to users across various fields in bioinformatics. Here’s a detailed breakdown of the FANTASIA pipeline, which consists of five key steps.
Step 1: Input File Preprocessing
The initial phase of the FANTASIA pipeline involves input file preprocessing, which is crucial for optimal performance. In FANTASIA v1, this step is mandatory. It eliminates redundant sequences, specifically those longer than 5,000 amino acids, ensuring efficient execution. Long sequences pose both computational and biological challenges due to disrupted signal representation when processed by certain models.
Conversely, FANTASIA v2 introduces more flexibility by making length filtering optional. Users can specify maximum sequence lengths and set a minimum sequence identity threshold, allowing for more tailored analyses based on their specific needs. For example, when benchmarking against closely related species, users may choose to filter out those sequences to enhance accuracy.
Step 2: Embedding Computation
Once the input sequences are prepped, the next step is embedding computation. FANTASIA excels in this area by providing protein embeddings that scale with proteome size. In FANTASIA v1, the pipeline supports the ProtT5 model popularized by Rostlab. However, FANTASIA v2 enhances these capabilities further by introducing support for ESM2 and ProstT5 models, enabling users to configure analyses according to their specific research questions.
Embeddings are generated through a series of commands that capture protein-level features efficiently. These embeddings are stored in HDF5 format for subsequent use in similarity calculations, retaining the integrity of the protein data across multiple models.
Step 3: Computation of Embedding Similarity
The core function of FANTASIA lies in calculating the distance between the input sequence embeddings and those stored in the reference database. FANTASIA v2 boasts an advanced structure that utilizes a PostgreSQL-managed reference vector database. Here, users can opt for either Euclidean distance or cosine similarity, each offering its unique benefits. While Euclidean distance aligns with traditional methods, cosine similarity captures directional relationships, which can be particularly advantageous when comparing embeddings generated by different models.
Step 4: GO Terms Transference
Following similarity calculations, the pipeline proceeds to GO terms transference. FANTASIA retrieves the closest protein embeddings, transferring their Gene Ontology (GO) terms to the input proteins based on defined parameters. Users can specify a distance threshold, allowing for granularity in analyses and tailoring results to their project’s requirements.
Both versions of FANTASIA utilize the latest GOA data for annotations, with FANTASIA v2 providing additional flexibility for employing older data versions if needed. This ensures that users are always working with the most relevant information, further bolstering the accuracy of functional annotations.
Step 5: Output Description and Formatting
The final step encompasses output description and formatting. This step provides users with a clear and organized view of the functional annotations assigned to their input proteins, complete with reliability indices. In FANTASIA v1, the output format is tab-separated, while v2 offers a more versatile comma-separated file (CSV) format that includes additional metadata, such as GO categories and protein language model (pLM) used.
This structured output not only enhances interpretability but also simplifies integration into broader workflows in functional genomics.
Computational Resources
The FANTASIA pipeline is designed to run efficiently on both CPUs and GPUs, although GPU utilization is recommended for speedier processing. The computational complexity remains manageable, scaling with the number of input sequences, which is a significant advantage for users handling large proteomics datasets.
Data Processing and Evaluation
FANTASIA has already been tested across diverse computing environments, showcasing its versatility in processing large-scale proteomic data. The system has been effectively employed to annotate extensive datasets, which cover nearly all animal phyla, allowing researchers to delve into functional genomics with newfound efficacy.
In addition to direct annotations, FANTASIA facilitates comparisons between enriched GO terms across different species, enabling researchers to visualize functional similarities and divergences across the animal kingdom. This brings a crucial interpretive layer to the data, enhancing the overall understanding of biological processes across various organisms.
Robustness and Flexibility in Research
With the wealth of functionalities provided by FANTASIA, researchers can optimize their functional annotation processes, ensuring that they can adapt methods to suit specific requirements. It empowers users to conduct comprehensive analyses while keeping user experience in mind.
FANTASIA is not just a tool; it is a gateway to accelerated discoveries in functional genomics, paving the way for deeper insights into the roles and functions of proteins across a vast array of species. As the field evolves, FANTASIA stands as a robust foundation for future innovations in bioinformatics and protein analysis.