Advancements in Single-Cell Foundation Models: A Detailed Examination
The emergence of large language models (LLMs) has captured the imagination of the scientific community, inspiring initiatives to harness similar foundational models within the realm of biology. The underlying premise is that if LLMs can successfully represent and manipulate vast amounts of textual knowledge, then analogous deep learning models could redefine our understanding of biological systems, particularly at the single-cell level. Recent studies have embarked on training foundational models based on extensive transcriptomic data from millions of individual cells, pushing the frontiers of genetic understanding and prediction.
Foundation Models in Biology
The idea of foundation models in biology is still in its nascent stages. A growing number of studies propose models like scGPT and scFoundation, specifically engineered to predict gene expression changes resulting from genetic perturbations. These models shine a spotlight on the potential of deep learning techniques to disentangle complex genetic interactions and unravel the nuanced behavior of cellular mechanisms. However, as these models compete to claim superiority over established techniques like GEARS and CPA (Cellular Perturbation Analysis), it becomes crucial to assess their efficacy in real-world applications.
Model Performance Benchmarking
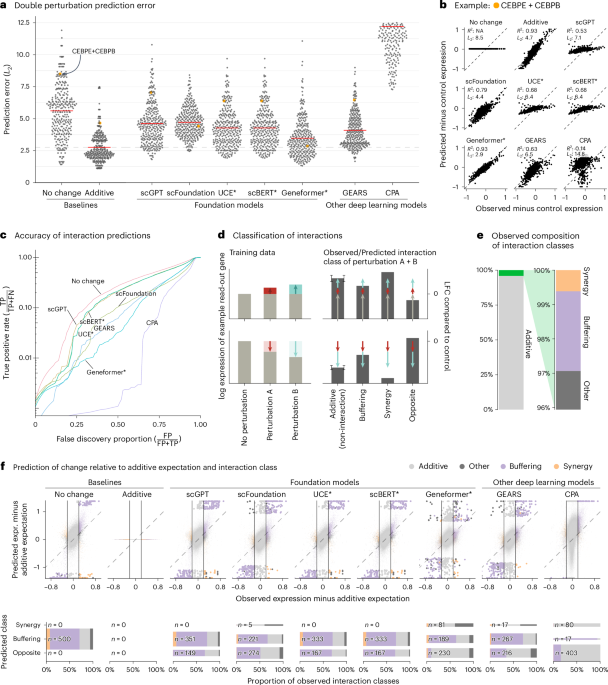
A comprehensive evaluation of the predictive capabilities of these models focused on double perturbations—situations where the expression levels of two genes are manipulated simultaneously. Leveraging well-curated datasets, researchers fine-tuned these models to assess errors in their predictions against observed expressions. Interestingly, the foundational models often exhibited higher prediction errors than even simplistic approaches that merely predicted no change or summed individual gene effects.
Detailed Implementation and Methodology
The methodology involved evaluating various models, including scBERT, Geneformer, and Universal Cell Embeddings (UCE), against more classical, simple baselines. Each model was subjected to rigorous testing across multiple random partitions of data to ensure robust validation. Parameters like the L2 distance between predicted and actual expression levels revealed that while some advanced models claimed intuitive insights into genetic interactions and perturbations, they frequently fell short of expectations. Surprisingly, many of these deep learning models—despite their complexity—did not outperform simpler statistical strategies employed in prior biological analyses.
Double Perturbation Prediction
In the context of double perturbations, the researchers employed phenotypic data arising from genetic interventions on K562 cells. The findings underscored that not only did the advanced models fail to yield better predictions than baseline methods, but they also struggled to capture the biological intricacies inherent in these multi-gene interactions. Detailed plots illustrated the substantial prediction errors across the board, drawing attention to the necessity for refinement in model architecture or training methodologies.
Interaction Predictions & Genetic Complexity
One fascinating aspect of genetic interactions is their unexpected nature. For instance, the necessity to identify when combined genetic manipulations yield results that are surprising—or distinctly different from what would be expected based on individual effects—proved to be a complex challenge. Despite the ambition of these more complex models, they frequently yielded a higher prevalence of buffering interactions, thereby minimizing the potential for genuinely synergistic outcomes.
Generalization to Unseen Perturbations
The ability of these models to predict effects from unseen perturbations is a particularly enticing feature. Models like GEARS leverage Gene Ontology annotations to extrapolate knowledge from existing data. In contrast, the foundation models claim to encapsulate genetic relationships inherently learned during their extensive pretraining phases. However, initial benchmarks indicate that, even though they hold promise, these models often do not outperform traditional statistical predictions, suggesting that their capacity for general applicability remains limited.
Simplicity vs. Complexity
A significant insight from the assessments is the continuing effectiveness of simpler linear models, especially when provided with appropriate pre-trained embeddings from gene and perturbation data. When embedding matrices were extracted from scFoundation and scGPT and integrated into simpler models, performance metrics often exceeded those of the more intricate models. This speaks volumes about the trade-offs inherent in model complexity versus predictive accuracy.
Limitations and Future Directions
Although the ongoing development of foundation models is riveting, there remain substantial limitations that need addressing. Current benchmarks rely heavily on datasets derived from cancer cell lines, which may not fully encapsulate the breadth of biological variance present in different tissues or organisms. Additionally, the existing models did not always manipulate data quality adequately, which might yield skewed results dependent on non-ideal workings of individual perturbations.
The Road Ahead in Perturbation Predictions
Deep learning continues to show promise across various domains, including single-cell genomics. However, as underscored by recent studies, the effective prediction of perturbation effects remains an uphill task. A concerted emphasis on meticulous benchmarking and performance assessment could lead to breakthroughs, ultimately paving the way for more robust applications of AI in individualized medicine and genetic research.
The insights derived from rigorous benchmarking of foundational models against simpler models illuminate both the exhilarating potential and the pressing challenges faced in this rapidly evolving field. Such evaluations not only foster scientific accountability but elevate the overall trajectory of research within biology, guiding future innovations in model design and validation.