Constructing Comprehensive Large-Scale Datasets for 26 Viral Families
In the realm of virology, understanding the relationship between viruses and their hosts is critical, particularly as we navigate a world increasingly challenged by zoonotic diseases. Our recent study delves into the construction of comprehensive datasets across 26 viral families, fundamental for predicting zoonotic potential—an essential pursuit in pandemic preparedness.
Building Tailored Models for Viral Families
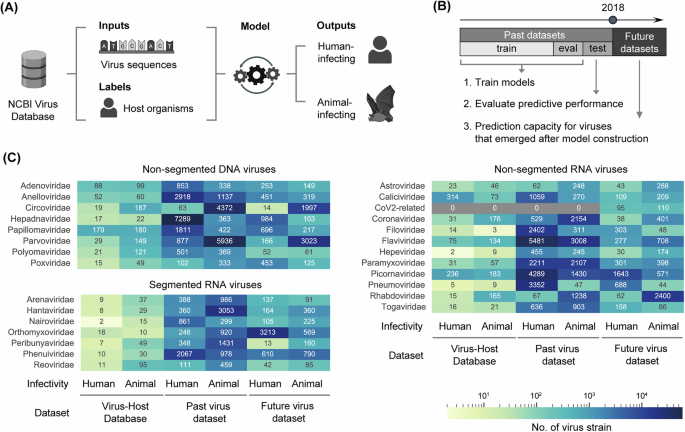
Our approach involved developing distinct models tailored to each viral family by training with paired viral sequences and corresponding host information. While utilizing the Virus-Host Database, a curated source established for examining such relationships, previous studies often faced limitations due to dataset composition. The 2019 version of this database featured 13,396 viral sequences, yet only 77.6% of these were linked to eukaryotic hosts. This skew could inflate model performance, as viruses like bacteriophages may lead to easier predictions that don’t reflect the reality of human-infecting pathogens.
Recognizing these shortcomings, we pivoted to gathering data specifically from the NCBI Virus Database. This allowed us to encompass viral families crucial for human health, including those with key pathogens. Our datasets drew from a vast array of 1,476 vertebrate and 535 arthropod species, significantly enhancing the available data.
Our meticulously curated datasets surpassed the Virus-Host Database by approximately 29 times, ensuring a more robust resource for developing predictive models aimed at identifying human-infecting viruses.
Evaluation Strategy and Model Development
In venture to bridge the knowledge gap and enhance predictive capabilities, we split our virus data into past datasets—comprising sequences identified until the end of 2017—and future datasets for assessing models against newly discovered viruses post-2018. This strategic division aimed to simulate real-world data availability and challenges.
Addressing the challenge of limited labeled data, we leveraged large language models (LLMs) pre-trained on extensive genetic sequences. Fine-tuning two pre-trained BERT models—DNABERT, tuned to the human genome, and ViBE, focused on viral genomes—became our pathway to extracting features associated with human infectivity. This innovative approach took into account molecular mimicry of host organisms, a known factor influencing a virus’s host range and potential for replication and immune evasion.
Performance Comparison: Insights from Past Virus Datasets
Our findings illuminate the effectiveness of our models when predicting viral infectivity utilizing past datasets. The benchmarking against existing models underscored that our BERT-infect pathways consistently outperformed other models across various viral families. A stark contrast was drawn with models lacking pre-trained weights, which faltered in predictive abilities. Precision-recall scoring underscored the disparity in model performance, advocating for the significant role of LLM pre-training, especially in contexts with limited labeled data.
Interestingly, our models impressed particularly in evaluating segmented RNA viruses—often overlooked in earlier assessments—highlighting their relevance in emerging infectious diseases. The performance metrics reveal a compelling picture of our datasets’ effectiveness and the predictive prowess of models built upon them.
Detecting Zoonotic Viruses: Practical Applications
Our study didn’t stop at theoretical models; we evaluated the applicability of our findings in real-world scenarios, particularly in detecting human-infecting viruses from high-throughput sequencing data. Both 250 bp single-ended reads and longer viral contigs were employed to mirror practical applications. Notably, models retained robustness in performance regardless of input length, distinguishing them as suitable tools for mining viral sequences in real time.
However, computational demands varied significantly among models. While deep learning models like BERT-infect demonstrated the capacity to parse shorter sequences efficiently, they required substantial computational power and time, creating a trade-off between efficiency and accessibility in high-throughput sequencing contexts.
Predictive Strengths and Limitations Against Future Viral Datasets
To ensure future preparedness for pandemics, models must predict the infectivity of newly identified viruses accurately. Our analysis explored various thresholds for evaluating this predictive capability, revealing comparable performances across models when assessing known data. However, challenges emerged in recognizing specific high-risk viruses, such as those related to SARS-CoV2, illuminating considerable gaps in current model frameworks.
Upon deeper inspection, certain viruses—especially H5 influenza A—were identified as high-risk yet inadequately flagged by models, revealing systemic weaknesses in our predictive abilities regarding zoonotic risks.
Challenges in Characterizing High-Risk Viral Lineages
Our analysis also unveiled significant challenges in predicting human infectivity across viral families characterized by evolving genetics. By mapping phylogenetic relationships, we identified species like Flavivirus, where frequent shifts in infectious potential complicated predictions. This complexity is emblematic of the broader challenges that models face as they attempt to grapple with rapidly changing viral genetics and the implications for human health.
The revelation that previously high-performing models struggled to recognize emerging zoonotic threats highlights a critical area for advancement. To foster better preparedness, there’s an urgent need for adaptive models capable of evolving alongside the viruses they aim to predict.
Conclusion
Navigating the intricate landscape of viral infectivity prediction underscores both the strides made in data collection and the hurdles still in play. As we forge ahead, this landscape of understanding remains a vital frontier in ensuring public health resilience against the perennial threat of emerging zoonotic diseases. Through continuous enhancements in our predictive models, the hope is to stay one step ahead in identifying, assessing, and mitigating viral threats that could lead to future pandemics.