Fly Strains and Their Role in Research
Introduction to Drosophila Strains
Drosophila melanogaster, often referred to as the fruit fly, is a staple organism in developmental biology studies. The versatility and genetic simplicity of these flies make them ideal models for investigating the genetic and molecular underpinnings of development. Among the various strains used, the Oregon-R (OreR) strain is commonly employed as the wild-type reference.
The Importance of Oregon-R Strain
The OreR strain offers a standard genetic background, enabling researchers to compare variances and derive insights into developmental processes. Understanding how different genetic factors influence development can lead to discoveries about fundamental biological principles applicable across species.
Types of Fly Strains Utilized
In addition to OreR, several specialized strains are used to visualize and track developmental processes in Drosophila embryos:
-
Histone H2A-RFP Strain (
his2av-mrfp1):- This strain helps in labeling nucleosomes within the embryo, offering real-time insights into nuclear dynamics and chromatin behavior. This strain can be obtained from the Bloomington Drosophila Stock Center (stocks 23650, 23651).
-
Live Imaging Strains:
- Strains like
yw; histone-rfp; mcp-nonls-gfpandyw; hb BAC>ms2were developed for studying specific activities, such ashb(hunchback) gene activity. These strains were generously provided by Dr. Hernan H. Garcia from the University of California at Berkeley.
- Strains like
- Bicoid Strain (
1×bcd):- Developed to assess the effects of Bicoid activity on early developmental processes, this strain was a gift from Dr. Jun Ma of Zhejiang University. Interestingly, the sex of embryos from these strains was not considered in studies due to the non-discriminative nature of early developmental stages.
Research involving these fruit flies adheres to strict ethical standards, as indicated by compliance with institutional and national policies, negating the need for additional ethical approvals.
Designing smFISH Probes
To visualize specific genetic activities, small fluorescent in situ hybridization probes (smFISH) are designed:
-
Probe Composition:
- Individual sets of DNA oligonucleotides are synthesized to be complementary to target transcripts. For instance, 48 probes target the
hbtranscript and 33 forKr. This precise design ensures robust hybridization and allows for effective visualization of nascent transcript activity.
- Individual sets of DNA oligonucleotides are synthesized to be complementary to target transcripts. For instance, 48 probes target the
- Conjugation Techniques:
- Probes targeting
Krare typically conjugated with tetramethylrhodamine (TAMRA), while those for thehbgene can either be coupled with Alexa Fluor™ 647 or TAMRA.
- Probes targeting
This approach equips researchers with crucial tools to study gene expression dynamics during embryonic development.
Live Imaging Techniques
Sample Preparation
For live imaging, embryos from the his2av-mrfp1 strain are collected directly. The process involves crossing female virgins from one strain with males of another to ensure a suitable genetic mix. The embryos are then treated to remove the chorion and mounted between a semi-permeable membrane and a coverslip for imaging.
- Imaging Conditions:
- Live embryos are embedded in Halocarbon 27 oil and imaged under specific conditions using Zeiss confocal microscopes. The imaging involves capturing sequences at different resolutions and intervals for well-rounded time-lapse studies of embryonic development.
Data Acquisition
The captured images provide insights into gene expression patterns and molecular interactions. Specialized imaging protocols, such as utilizing 488 nm and 561 nm excitation wavelengths, allow for high fidelity in capturing distinct fluorescent signals from different genetic markers.
Fixed Imaging and Data Acquisition
To complement live imaging, fixed embryos are also analyzed. Embryos undergo treatment with paraformaldehyde for fixation, followed by immunostaining. This multi-pronged approach ensures both temporal and spatial aspects of genetic expression are captured.
- Immunofluorescence:
- Following fixation, specific proteins can be visualized through immunostaining techniques. This method enables the comparison of mRNA distribution with protein concentrations, providing a holistic view of regulatory networks in action.
Image Processing and Segmentation
The imaging data undergoes extensive processing to ensure accurate interpretation:
-
Nuclear Segmentation:
- Specialized algorithms like Cellpose can be employed for three-dimensional segmentation, allowing for precise counting and localization of nuclei within the embryo.
- Quantification:
- By identifying local maxima in fluorescence intensity, researchers estimate mRNA production. This enables them to extract key dynamics of gene expression, linking developmental timing to molecular activity.
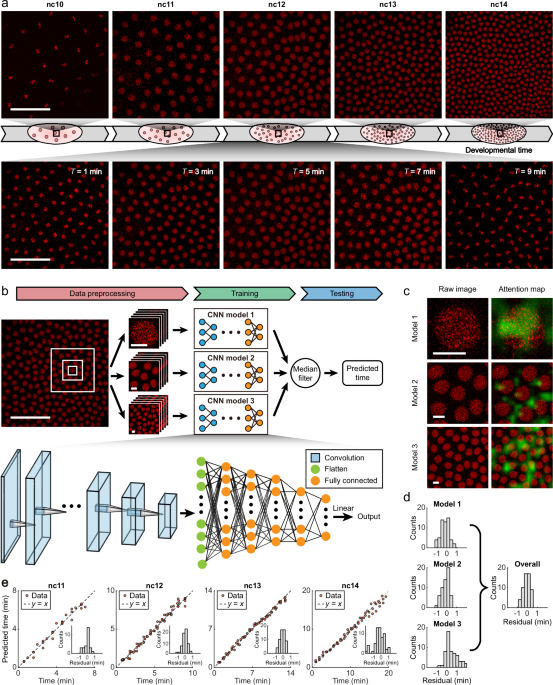
Advanced Computational Modeling
A significant aspect of modern genetic research is leveraging computational models to understand complex data patterns:
-
Machine Learning Applications:
- Deep learning models are developed to predict developmental timelines based on imaging data. These models integrate multiple datasets, refining predictions through k-fold cross-validation techniques, ensuring robust output from live and fixed image data.
- Statistical Approaches:
- Advanced statistical methods are used to measure nuclear size and dynamics, offering vital insights into the physical changes embryos undergo throughout development.
Integrating Data for Insights
Through the collected imaging data and developed models, researchers can integrate different datasets to generate comprehensive biological insights. For instance, comparing predictions from multiple imaging techniques allows scientists to assess the precision of their developmental timings and gene expression quantifications.
Overall, the advancements in floral imaging and computational biology represent a transformative period in developmental genetics, increasing our understanding of fundamental biological processes and their implications for broader biological functions. The manipulation and utilization of various Drosophila strains, paired with sophisticated imaging and quantitative analysis, continue to illuminate the intricate tapestry of life at the molecular level.