Advancements in Biomedical Image Segmentation: The Multimodality Cell Segmentation Challenge
Biomedical image segmentation is pivotal in fields like medical imaging, requiring precise identification and delineation of cellular structures from complex datasets. The latest research has highlighted significant strides through collaborative efforts aimed at addressing these challenges. A landmark paper titled "The multimodality cell segmentation challenge: toward universal solutions" by Ma et al. provides insights into current methodologies and the advancements achieved in this field.
The Multimodality Cell Segmentation Challenge
Ma et al. (2024) describe a prominent challenge that focuses on developing universal solutions for cell segmentation across multiple imaging modalities, including fluorescence, bright-field, and phase-contrast microscopy. This challenge is particularly significant as it addresses the need for efficient algorithms that can generalize across various imaging conditions and cell types.
The paper underscores the implications of such advancements on downstream applications, such as drug discovery, pathology, and cellular biology research. With a clear objective to bridge the gap between different imaging modalities, the researchers have established metrics and benchmarks to evaluate the performance of various segmentation algorithms.
You can read the full article here.
nnU-Net: A Step Forward in Deep Learning for Biomedical Segmentation
In the quest for effective segmentation techniques, Isensee et al. (2021) introduced nnU-Net, a self-configuring framework that adapts to multiple biomedical image segmentation tasks. This method automates the configuration process for U-Net architectures, significantly improving performance across various datasets.
The concept of nnU-Net revolves around leveraging transfer learning to enhance the model’s adaptability, reducing the need for extensive manual tuning. This adaptability is especially useful in biomedical contexts, where variability in data can complicate the training process. The nnU-Net framework represents a groundbreaking step in the evolution of segmentation techniques by minimizing the barriers to entry for researchers.
You can find more details in their publication here.
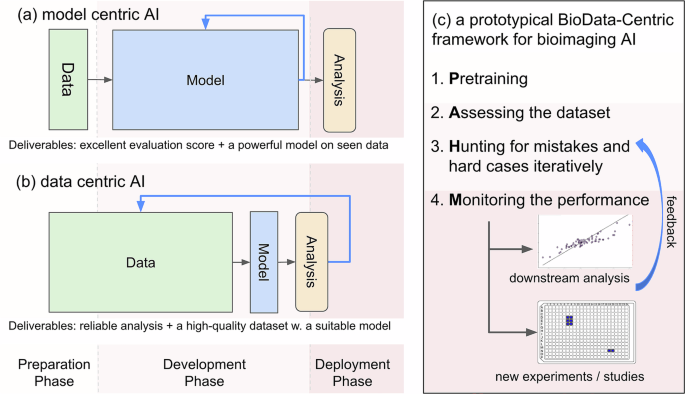
Data-Centric Approaches in Artificial Intelligence
Zha et al. (2023) provide a comprehensive survey on data-centric artificial intelligence, emphasizing the importance of high-quality training data in improving model performance. Their findings align with the theme of the multiclass cell segmentation challenge, highlighting how refining datasets enhances segmentation outcomes.
Essentially, a robust dataset allows models to better learn relevant features without overfitting. Techniques like active learning and data augmentation are critical in ensuring the continued relevance of datasets in the ever-evolving landscape of biomedical research.
To explore this further, you can view the complete survey here.
Continual Lifelong Learning in Neural Networks
Parisi et al. (2019) explore the paradigm of continual lifelong learning (CLL) within neural networks, addressing the challenges posed by catastrophic forgetting. In the context of biomedical image analysis, CLL can help models adapt to new data while retaining learned knowledge.
The significance of this approach becomes apparent in the biomedical field, where datasets frequently evolve as new research developments come to light. By implementing CLL mechanisms, models can maintain accuracy and relevance without being retrained from scratch, thus optimizing resources and time.
Check out their paper here for an in-depth look.
Addressing Uncertainty in Deep Neural Networks
Understanding the uncertainty associated with predictions in deep learning is critical. Gal and Ghahramani (2016) proposed using dropout methods as a Bayesian approximation to represent model uncertainty. This concept is particularly relevant in scenarios where precise segmentation is crucial, such as in identifying cancerous cells.
By incorporating uncertainties into segmentation frameworks, researchers can make more informed decisions about the reliability of the predictions, allowing for better clinical outcomes. The implications of such methodologies extend beyond segmentation, influencing the broader scope of predictive analytics in medicine.
For further insights, refer to their work here.
Label-Efficient Semantic Segmentation
The work of Zhou et al. (2021) presents an innovative approach to semantic segmentation through weak supervision, exploring how leveraging minimal labeled data can lead to effective segmentation outcomes in complex images. This is especially pertinent in biomedical applications, where obtaining labeled data can be labor-intensive and costly.
The essence of their method highlights a growing trend in machine learning: maximizing the utility of available data while minimizing the reliance on heavily labeled datasets. This approach promises to democratize access to advanced bioimage analysis tools across varying research environments.
Explore their findings in the publication here.
Sample and Data Efficiency Strategies
In contexts where labeled data is scarce, innovative strategies like data distillation and sample efficiency have emerged. Sachdeva and McAuley (2024) review data distillation techniques that optimize the relationship between model performance and the size of training data, which can be particularly beneficial in fields requiring rapid adaptation.
Such strategies are crucial for researchers who need to implement machine learning techniques in clinical settings efficiently. Optimizing how data is used helps maintain high performance while controlling costs.
For more on this topic, you can review their survey here.
Evaluation Without Ground Truth
A significant challenge in biomedical image segmentation lies in evaluating the performance of segmentation algorithms without ground truth data. This addresses a critical bottleneck in practical settings where manually annotating data can be impractical or impossible.
Recent works, such as that of Sims et al. (2023), explore novel metrics and methodologies for assessing segmentation quality in the absence of ground truth, opening avenues for robust algorithmic evaluation that supplements existing practices.
For detailed insights, check their work here.
Conclusion
The advancements in cell segmentation through multimodality approaches, enhanced algorithms, and efficient data utilization paint a promising picture for the future of biomedical imaging. As researchers continue to push the boundaries in this field, the integration of innovative techniques and methodologies will undoubtedly enhance our capabilities in accurately interpreting biological data, ultimately leading to better healthcare solutions.
Exploring and adopting these recent advancements can optimize both research outcomes and clinical applications, fostering a healthier future.